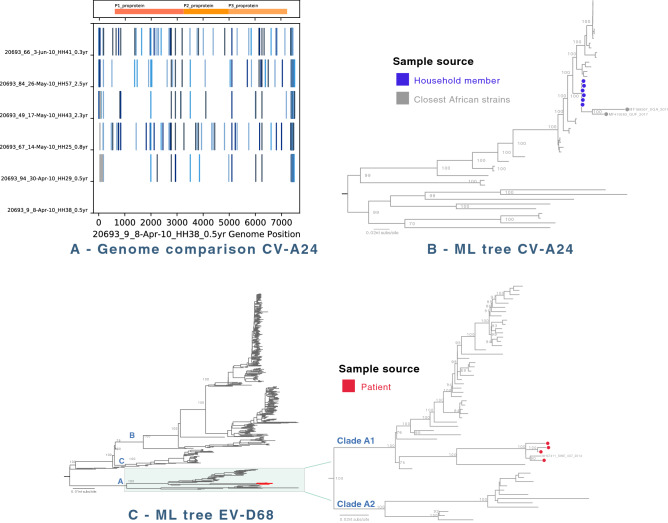
Figure 3.
(A) Genome comparison of identified human coxsackievirus A24 (CV-A24) from the study. Kilifi CV-A24 genomes were examined and compared against the earliest Kilifi CV-A24 genome identified (20693_9, sample collected on 8 April 2010). For each genome, nucleotide changes from the first CV-A24 genome (20693_3) were indicated by vertical blue lines, gaps in the sequence were indicated by grey bars. (B) Maximum-likelihood phylogenetic tree of CV-A24 genomes. The ML tree compared 6 Kilifi CV-A24 genomes (all from household cohort, indicated as blue circles) to global genomes. The tree was mid-point rooted for clarity and horizontal branch lengths were drawn to the scale of nucleotide substitutions per site, and significant bootstrap values were shown for major nodes. (C) Human enterovirus D68 (EV-D68) maximum-likelihood phylogenetic tree. Maximum-likelihood phylogenetic tree were inferred comparing 4 Kilifi EV-D68 genomes (all from KCH pneumonia patients cohort, highlighted in red) to global genomes. The tree was mid-point rooted for clarity and horizontal branch lengths were drawn to the scale of nucleotide substitutions per site, and significant bootstrap values were shown for major nodes. These four local EV-D68 viruses belonged to clade A1, as shown in the zoomed out tree.