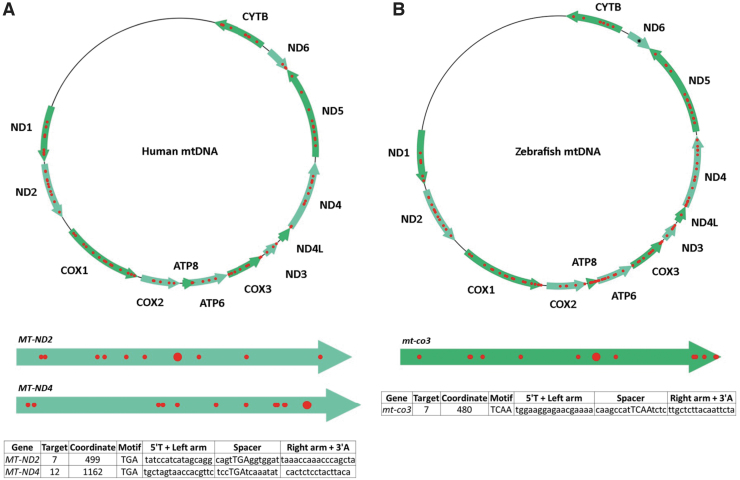
FIG. 2.
Targetable sites for PTC induction in human and zebrafish mtDNA. (A) Based on design parameters extrapolated from TALEN and initial mtDNA base editor work, and including assembly parameters for the FusX system, TALE Writer identified a total of 1617 potential target sites for C-to-T base editing in the human mitochondrial genome, 94 of which are amenable to PTC induction (red circles). The human mitochondrial genes MT-ND2 and MT-ND4 are represented below the human mtDNA map as light green arrows. The big red circles correspond to the targets chosen for this study. The table at the bottom includes the number of the targets within their respective genes, their local coordinates, their sequence motifs, and the sequences of their TALE arms and spacers. (B) Similarly, the zebrafish mitochondrial genome possesses a total of 1570 potential target sites for C-to-T base editing, 115 of which correspond to targets amenable to PTC induction (red circles). The zebrafish mitochondrial gene mt-co3 is represented below the zebrafish mtDNA map as a green arrow. The big red circle corresponds to the target chosen for this study. The table at the bottom includes the number of the target within mt-co3, its local coordinate, its sequence motif, and the sequences of its TALE arms and spacer. The single black asterisk in the zebrafish gene mt-nd6 represents a site for PTC induction made available by setting the maximum TALE arm and spacer lengths to 17 bp; such a FusXTBE format can be assembled via our FusX extender kit. mtDNA, mitochondrial DNA. PTC, premature termination codon. Color images are available online.