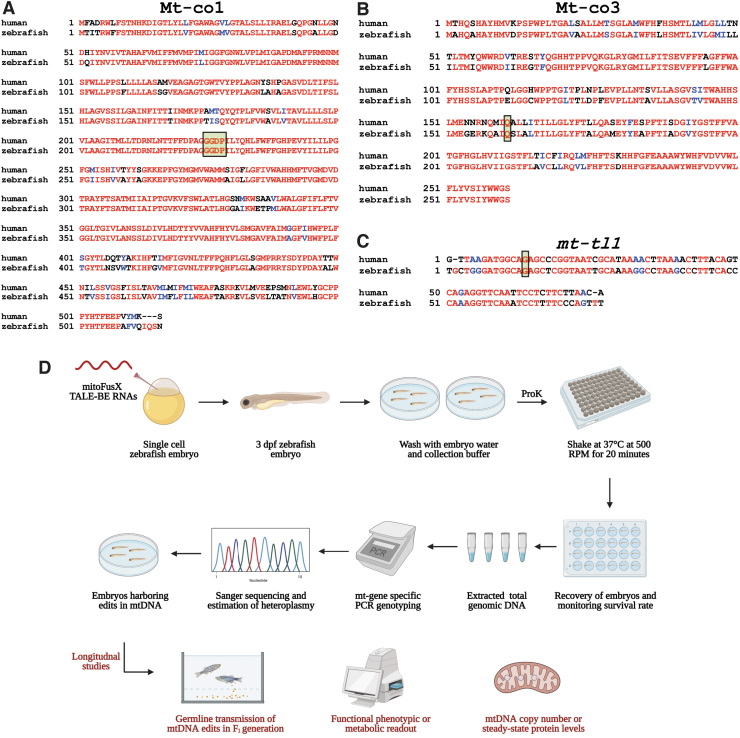
FIG. 5.
Schematic to prioritize base editing targets and noninvasively genotype mtDNA from zebrafish embryos. (A, B) Targets identified in the mitochondrial protein-coding genes, mt-co1 and mt-co3, were prioritized to test the editing efficiency of FusXTBE in vivo. Amino acid alignments are shown for the human MT-CO3 and MT-CO1 proteins with zebrafish Mt-co1 and Mt-co3 proteins. Edited amino acids are shaded and boxed. (C) The mitochondrial-encoded tRNA gene, mt-tl1, was prioritized to introduce base edits. Sequence alignment of human MT-TL1 and zebrafish ortholog mt-tl1 is shown. Target nucleotide is shaded and boxed. Red indicates sequences fully conserved between the two species. Blue indicates similar and black indicates positions with no similar or identical relationship. (D) Longitudinal study design assay for potential functional phenotype of the engineered edits in the zebrafish mtDNA locus. Zebrafish embryos are gently shaken in the genotyping buffer supplemented with proteinase K. Following recovery of the embryos, DNA is isolated from the shed cells, and mitochondrial genotyping is conducted for the target locus. Positive embryos can then be screened for germ line transmission or different functional and molecular studies. For Figure 4A–C, sequences were aligned using T-COFFEE multiple sequence alignment webserver and color-coded using BoxShade server. Figure 4D was created using Biorender.com. Color images are available online.