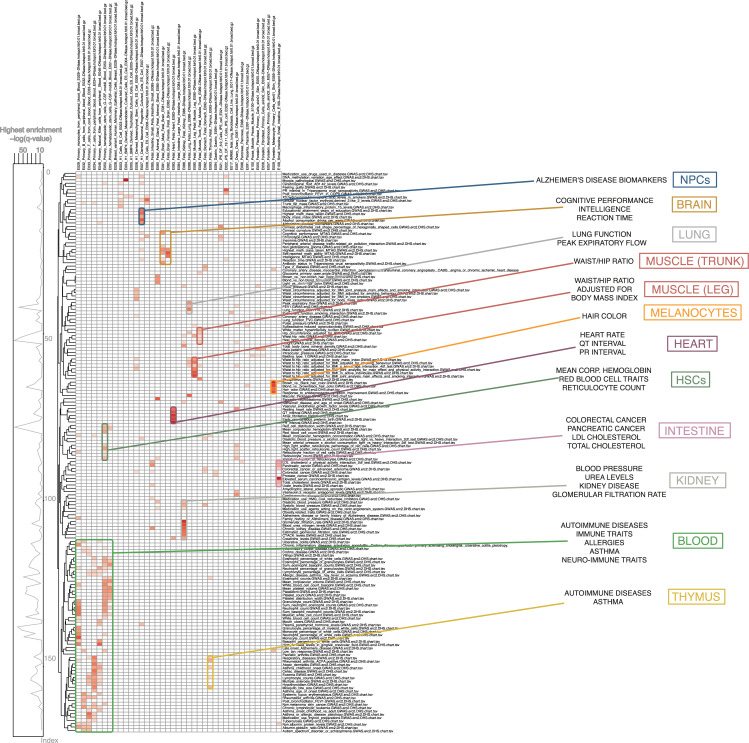
Fig. 1.
GWAS enrichments for DNase I hotspots from consolidated Roadmap Epigenomics consortium data: Shown are significant tissue-specific enrichment results (q value < 0.01, Benjamini-Hochberg (BH) correction) for FORGE2 analysis across the NHGRI/EBI GWAS catalogue (downloaded 2 September 2019). GWAS phenotypes (rows) are clustered using complete linkage clustering (Euclidean distance), while different tissues and cell types (columns) are grouped according to related tissue or cell type categories (e.g. white blood cell categories are placed together, as are fibroblast categories, etc.). Values are row-normalised, with reference lineplot on the left indicating top enrichment value for each category. Clustering reveals groups of related phenotypes with similar tissue-specific enrichment profiles, such as immune traits for blood or cognitive performance for brain. Groups are highlighted for different tissue- and cell type-enrichments, each in a different colour (right panel). A zoomable version of this figure can be found at: https://www.easyzoom.com/imageaccess/797f4272e4674bb28b42adf3f1918cf7