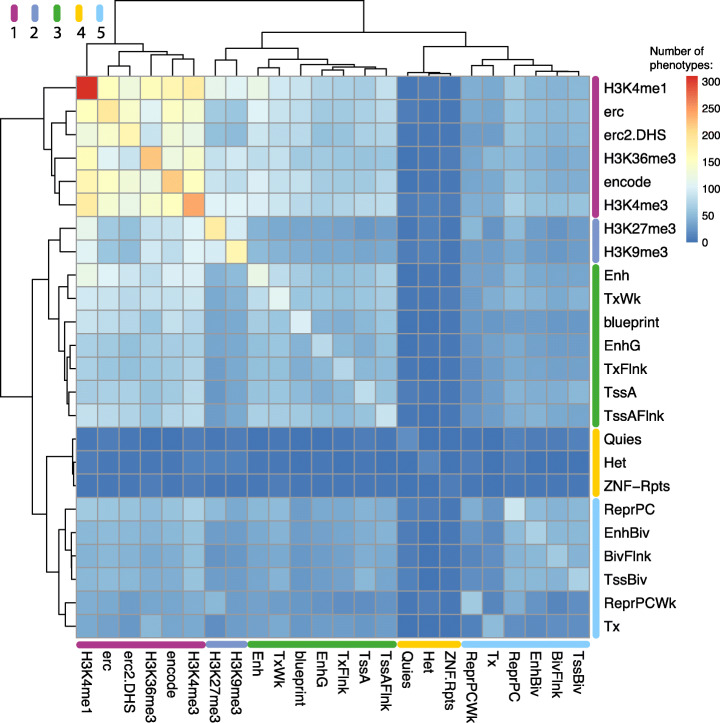
Fig. 5.
Relationship between enriched GWAS phenotypes for different epigenomic marks from consolidated Roadmap Epigenomics consortium data: heatmap showing the number of tissue-specific enriched phenotypes (q value < 0.01, BH correction) for FORGE2 analysis across the GWAS catalogue for DNase I hotspots, histone mark broadPeaks and HMM chromatin states. Epigenomic mark categories are clustered using complete linkage clustering (Euclidean distance), across rows and columns. Clustering reveals 5 main groups of epigenomic marks, coloured in purple (group 1), dark blue (group 2), green (group 3), yellow (group 4), and light blue (group 5). A zoomable version of this figure can be found at: https://www.easyzoom.com/imageaccess/4e114698194f492faa1d6812d325cd30