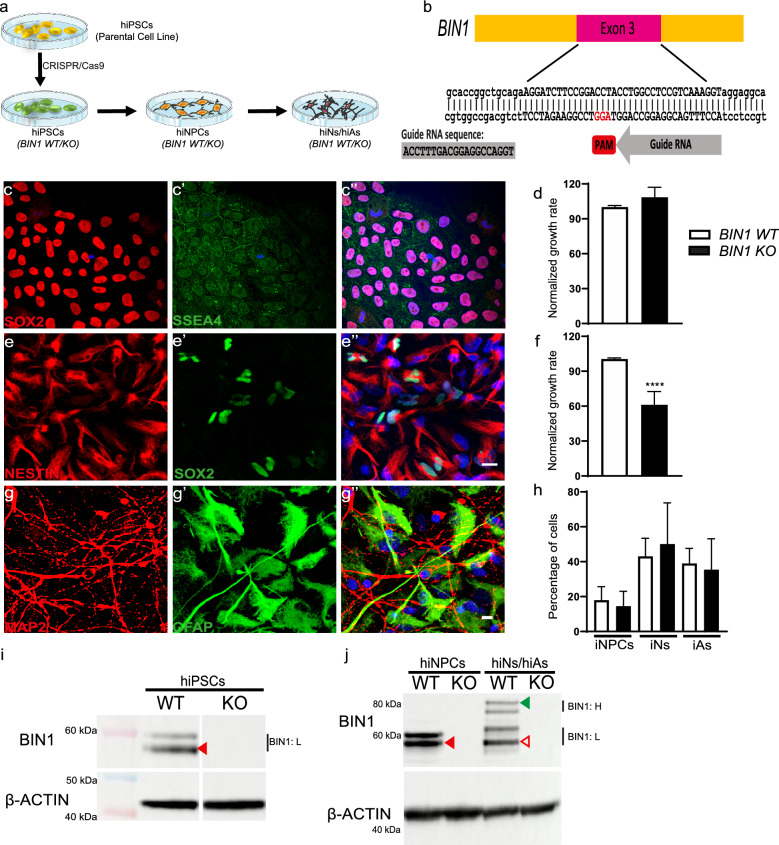
Fig. 5.
Characterization of hiPSCs and their cell derivatives. a Schematic showing the production of BIN1 WT and KO hiPSCs from parental cell line using CRISPR/Cas9 technology. These hiPSCs, in turn, were used to generate intermediate hiNPCs, and subsequently, mixed cultures of hiNs and hiAs. b Representation of exon 3 region of BIN1 was targeted for the production of BIN1 WT and KO hiPSCs by CRISPR/Cas9 technology. The guide RNA sequence is shown (grey). c-c’’ Representative images showing pluripotency markers SOX2 (red), SSEA4 (green) and stained with Hoechst 33258 (c’’) in BIN1 WT and KO hiPSCs. d Plot showing the normalized growth rate of BIN1 WT and KO hiPSCs (N = 4 independent cell passages; p = 0.77, Unpaired t-test). e-e’’ Representative images showing hiNPCs labelled for NESTIN (red), SOX2 (green) and stained with Hoechst 33258 (e’’, Scale Bar = 20 µm). f Plot showing the normalized growth rate of BIN1 WT and KO hiNPCs (N = 9 independent cell passages: ****p < 0.0001, Unpaired t-test). g-g’’ Representative images showing a 6-week-old mixed hiNs/hiAs culture labelled for the neuronal marker MAP2 (red), the astrocytic marker GFAP (green) and stained with Hoescht 33258 (g’’, Scale Bar = 10 µm) h Plot showing the percentage of cells in different cell populations – hiNPCs, hiNs, and hiAs (ANOVA F(5,12), p = 0.45). i Immunoblot for BIN1 and actin in BIN1 WT and KO hiPSCs samples. Band indicated on the blot for the WT cells (solid red arrowhead) indicates BIN1-light (BIN1:L) isoforms of BIN1. j Immunoblot showing the expression of BIN1:L isoforms (solid red arrowhead) in WT hiNPCs and both BIN1:L (hollow red arrowhead) and BIN1-heavy (BIN1:H; solid green arowhead) isoforms in 6-week-old mixed cultures of hiNs and hiAs