FIGURE 1.
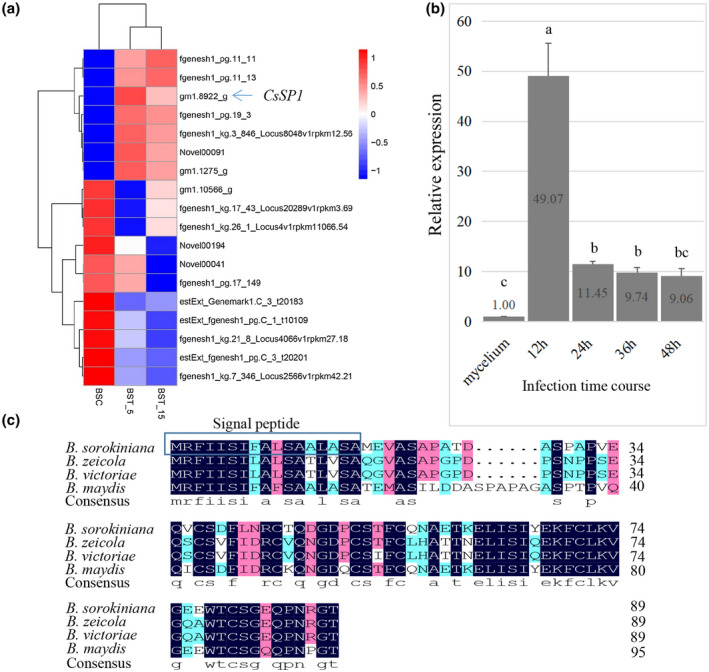
Differentially expressed genes (DEGs) in the transcriptome and CsSP1 feature analysis. (a) Cluster analysis of DEGs from the wheat Aikang 58 cultivar infected with Bipolaris sorokiniana. (b) Reverse transcription quantitative PCR (RT‐qPCR) confirmation of the expression pattern of CsSP1 in planta. Aikang 58 leaves were inoculated with 105 spores/ml of B. sorokiniana Lankao 9‐3. The inoculated leaves were placed in a moist chamber in the dark for 24 h and then kept in a greenhouse at 25°C (47% humidity) with a 16 h light/8 h dark photoperiod. The leaves were sampled at 12, 24, 36, and 48 h for total RNA extraction, while mycelia cultured for 2 days in YEPD were used for fungal RNA extraction. The experiments were repeated three times. The expression levels were calculated using the 2−∆∆ C t. Significant differences calculated by Tukey's LSD, p < 0.05. (c) Alignment of CsSp1 orthologues from Bipolaris spp. was performed using DNAMAN. BSC: B. sorokiniana mycelia from liquid medium served as controls. BST_5 and BST_15: assays of Aikang 58 root and stem base samples from plants growing in pots at 5 and 15 days after B. sorokiniana infection. Signal peptides were predicted by SignalP v. 4.0 software (http://www.cbs.dtu.dk/services/SignalP/)
