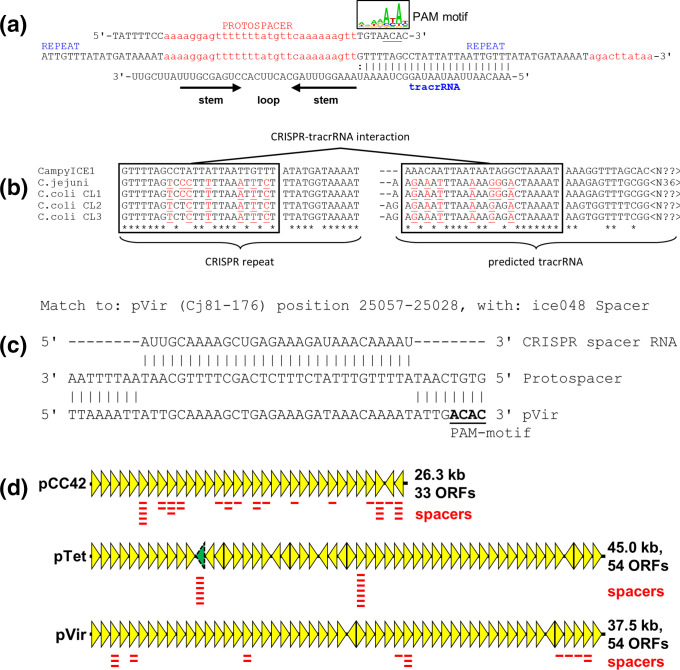
Fig. 5.
Characteristics of the CampyICE1 CRISPR spacers, protospacers and tracrRNA, and predicted plasmid targeting by the CampyICE1 CRISPR-Cas9 system. (a) A section of the CRISPR array is shown (centre) with the corresponding protospacer (top) with 8 nt flanking sequences which contain the PAM motif at the 3′ end of the protospacer, represented using a sequence logo. The tracrRNA sequence and structure are included below. (b) Comparison of the CRISPR-repeats and predicted tracrRNA part of CampyICE1, C. jejuni and the three C. coli clades. The tracrRNA and CRISPR-repeat show matching changes as indicated by red underlined residues. Asterisks indicate conserved nucleotides, boxes indicate the complementary sequences in CRISPR repeat and tracrRNA. (c) Example of a CampyICE1 CRISPR spacer perfectly matching a segment of the C. jejuni 81-176 pVir plasmid. (d) Schematic representation of the pCC42, pTet and pVir family of plasmids (based on the C. coli 15-537360 pCC42 plasmid and the C. jejuni 81-176 pTet and pVir plasmids), with the locations of plasmid-targeting CampyICE1 spacers indicated. For pTet, the approximate location of target gene YSU_08860 (absent from the C. jejuni 81-176 plasmid) is indicated by the dashed, green arrowhead. More information on specific spacers and their targets is provided in Table S2.