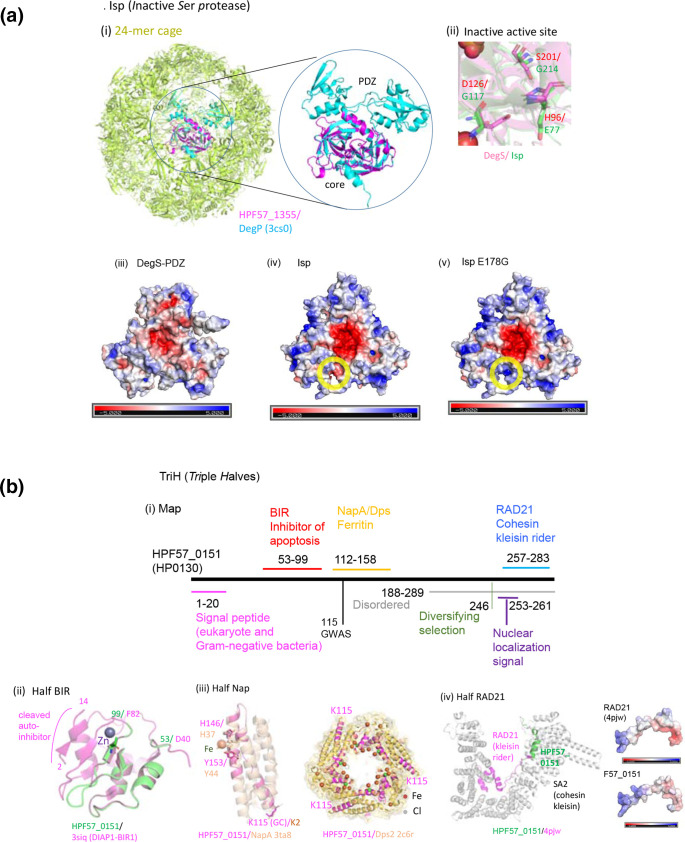
Fig. 5.
Predicted structures of three new virulence factor/oncoprotein candidates. (a) Isp (inactive Ser protease). (i) F57_1355 modelled on E. coli DegP. (ii) Active site of F57_1355 modelled on 3lgi.1 in PDB (E. coli DegS). The three amino acids (HDS triad) responsible for activity are all replaced. (iii)–(v) Surface electric charge distribution in E. coli DegS without PDZ [75] (3lgi.1 in PDB), HPF57_1355 modelled on it, and the E178G mutant generated in silico. (b) TriH, Triple halves. HPF57_0151 (HP0130). (i) Map. ‘Disordered’ is from UniProt. Nuclear localization signal is by cNLS Mapper. ‘Diversifying selection’ is from a previous study [49]. (ii)–(iv) Similarity to three half domains. (iii) Modelled on NapA (strain YS39, 4evd in PDB) and aligned with iron-soaked NapA (YS39, 3ta8 in PDB). Fe-interacting residues as well as the GWAS residues are in sticks. The 2c6r in PDB is Dps2 in Deinococcus radiodurans . Note the difference in NapA coordinates in the literature [48, 76]. (iv) HPF57_0151 modelled on PDB 4pjw (human).