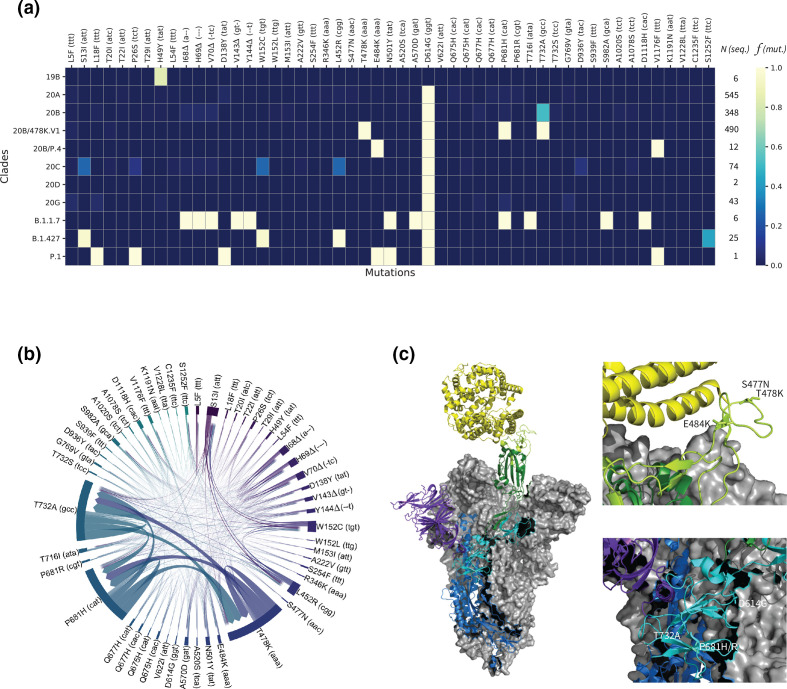
Fig. 2.
Systematic sequence/structure analysis of the spike protein’s mutations of concern. (a) Heatmap highlighting relative frequencies f for 51 important mutations (out of 315) across 1552 viral genomes arranged in 11 clades (19B, 20A–D, 20B/478 K.V1, 20B/P.4, 20G, B.1.1.7, B.1.427, P.1). N indicates the number of available genome sequences per clade. See Fig. S5 for a heatmap including all mutations. (b) Chord diagram displaying the covariances between the 51 most frequent mutations. The thickness of the arrows denotes the strength of a covariance. (c) Left: electron microscopy structure (PDB:7A94 ref) of the trimeric spike (S) glycoprotein of SARS-CoV-2 bound to one angiotensin-converting ewnzyme-2 (ACE2, yellow). Two chains of the viral protein are shown in grey surfaces. The third chain comprises the N-terminal domain (purple), the S1 (cyan) and S2 (blue) protein subdomains, the receptor-binding domain (RBD, green), and the receptor-binding motif (RBM, light green). Right: positions of the important mutations S477N, T478K, E484K, D614G, P681H/R and T732A in two different regions.