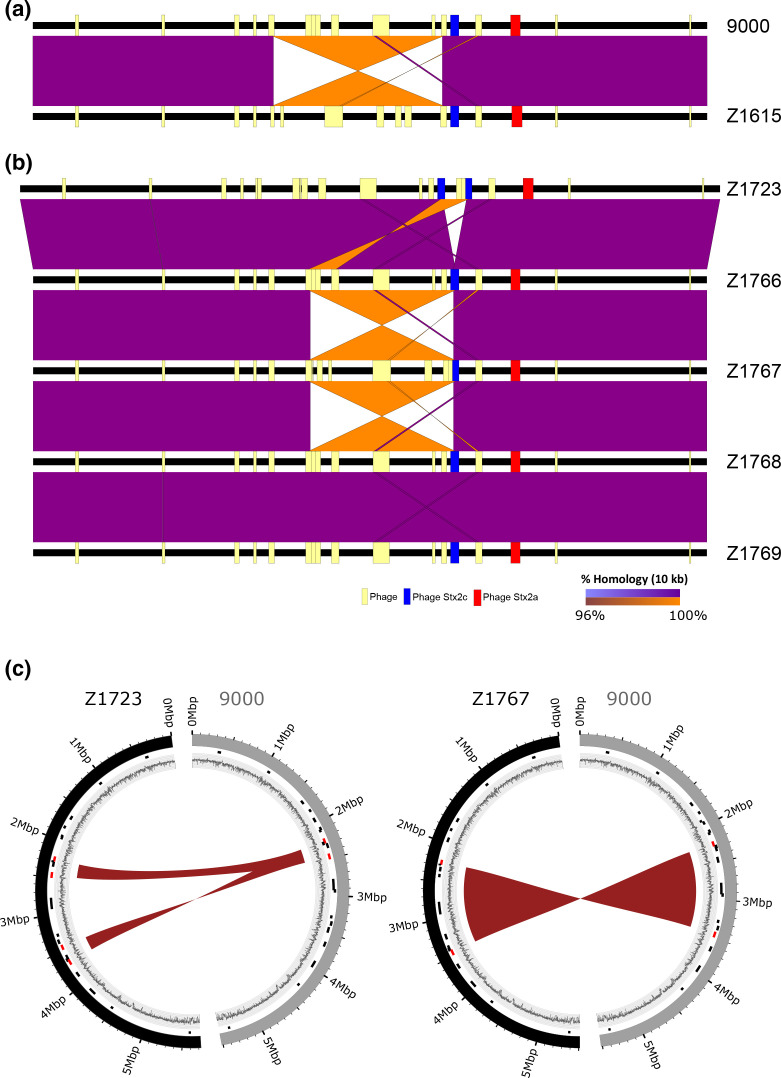
Fig. 4.
Detection of LCRs in E. coli O157:H7 PT21/28 strain 9000 variants analysed from cattle colonization studies. Pairwise whole-genome comparisons of strains from trial 1 (a) and trial 2 (b) are shown with direct (purple) and inverted (orange) homology at a blast cut-off of 10000 bp between strains. Whole genomes (black lines) are centred by the replication terminus (Ter) and the loci of prophage (yellow boxes) and Stx prophage (ΦStx2c;blue and ΦStx2a;red) in each strain are mapped. (c) Circos plots showing the identified 220 kbp duplication in Z1723 (left) and 1.2 Mbp inversion in Z1767 (right) relative to progenitor strain 9000. Outer ring: strain 9000 (grey) and LCR derivatives Z1723 and Z1767 (black); middle ring: loci of prophage (black) and prophage a LCR boundaries (red); inner ring: GC content.