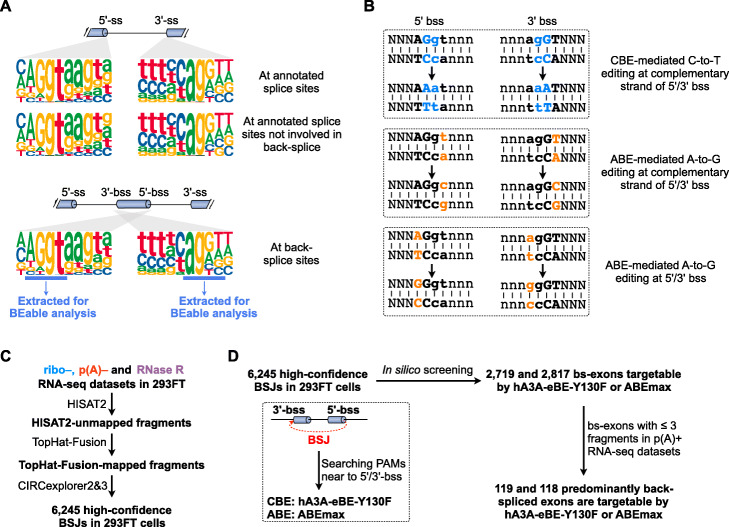
Fig. 1.
Consensus sequence analysis of (back-)splice sites and prediction of their availability to base editing. A Genome-wide analyses of consensus sequences at all 5′/3′ splice sites (5′/3′ ss), 5′/3′ ss without back-splice (top), or 5′/3′ back-splice sites (5′/3′ bss) (bottom) of annotated exons. Ten bases around 5′ bss/5′ ss (three upstream exonic bases and seven downstream intronic bases) and ten bases around 3′ bss/3′ ss (seven upstream intronic bases and three downstream exonic bases) were fetched for consensus sequence analysis. Intronic sequences were represented by a, t, c, and g, and exonic sequences were represented by A, T, C, and G. B Diagram of directing base editor (BE) to introduce base mutation at 5′/3′ bss. CBE could lead to C-to-T base editing at complementary strands of 5′/3′ bss. ABEs could introduce A-to-G base editing at 5′/3′ bss or at their complementary strands. C Prediction of circRNAs from ribo−, p(A)−, and RNaseR RNA-seq datasets from 293FT. D In silico screening of circRNAs with predominantly back-spliced exons could be targeted by hA3A-eBE-Y130F or ABEmax at back-splice sites