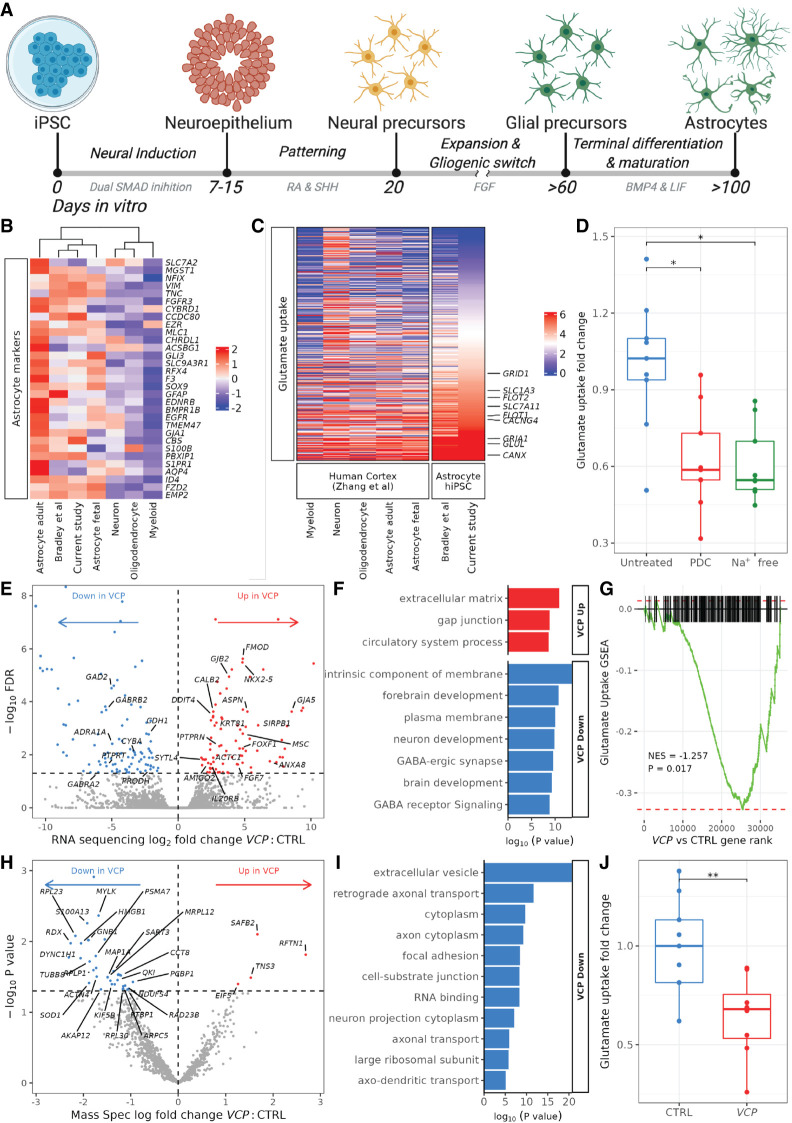
Figure 1.
Characterization of control and VCP-mutant hiPSC-astrocyte cultures. (A) Schematic showing directed differentiation of hiPSC-astrocytes patterned to the ventral spinal cord. (B,C) Heatmaps showing the mean row-scaled variance stabilized gene counts of astrocyte-specific markers (B) and glutamate uptake genes (C) in our control hiPSC-astrocytes (current study), ventral spinal cord (VSC) regional hiPSC-astrocytes (Bradley et al. 2019), and human CNS tissue purified cell types (Zhang et al. 2016). For individual sample clustering, see Supplemental Figure S1. (D) Boxplot showing fold change in glutamate uptake of control hiPSC-astrocytes treated with 1 mM PDC or incubated in Na+-free media over untreated. N = 5 control lines each with one to three assays (experimental repeats) and each with one to three wells on the plate (technical replicates). A Wilcoxon test was used to compare groups (PDC vs. untreated, P = 0.012; Na+ free vs. untreated, P = 0.012). (E) Volcano plot showing log2 fold change in VCP-mutant versus control astrocyte differential gene expression from RNA sequencing. Genes with significantly (FDR < 0.05) increased expression are shown in red, and those decreased in expression are shown in blue. (F) Gene Ontology (GO) terms enriched in up-regulated (red) and down-regulated (blue) VCP versus control astrocyte differentially expressed genes from RNA sequencing. (G) Gene set enrichment plots showing changes in gene expression with VCP versus control astrocytes for the glutamate uptake gene set. (NES) Normalized enrichment score. (H) Volcano plot showing log fold change in VCP-mutant versus control astrocyte differential protein abundance from mass spectroscopy. (I) GO terms enriched in down-regulated differential proteins from mass spectroscopy in VCP-mutant versus control astrocytes (there were no terms associated with up-regulated proteins). (J) Fold change in glutamate uptake of control (blue) and VCP-mutant (red) astrocytes normalized to confluency. N = 5 control lines, four VCP lines, three technical replicates, one to four experimental repeats. To normalize for variation between the four experimental repeats, fold changes were calculated by dividing the mean of the three technical repeats over the control samples for each experimental repeat. Wilcoxon test: (**) P < 0.01.