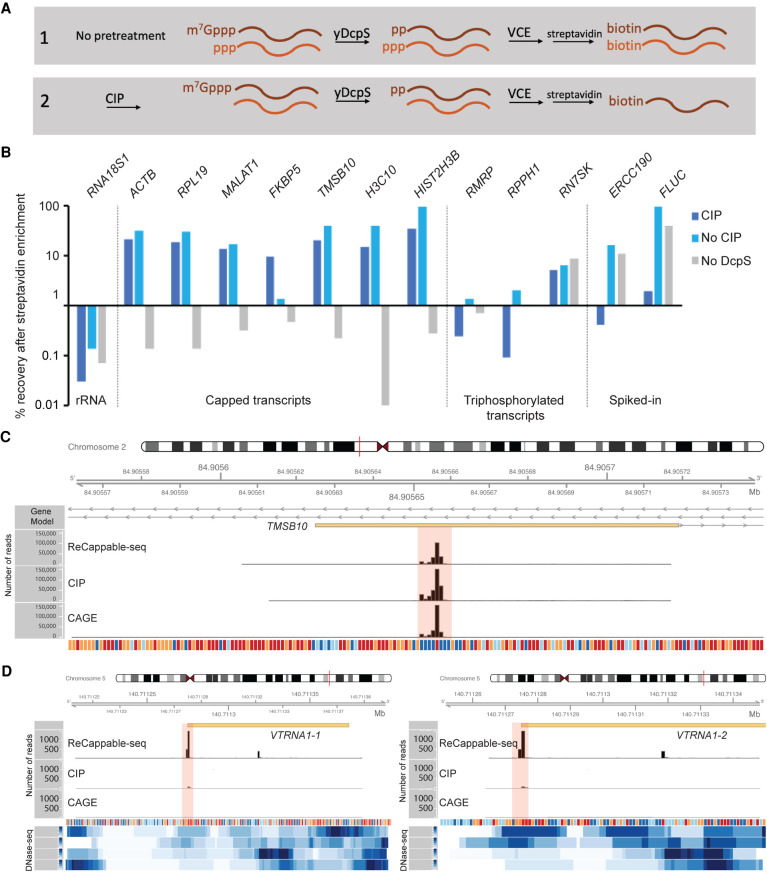
Figure 1.
ReCappable-seq. (A) Principle of ReCappable-seq. (1) RNA is subjected to decapping with yDcpS, which acts on capped transcripts originating from Pol II transcription. Subsequently, the RNA is capped with a biotin-modified GTP analog (3′-desthiobiotin-GTP) using VCE. This biotinylation step allows enrichment of all primary transcripts on a streptavidin matrix. (2) Differentiation of Pol II from the non–Pol II transcripts is accomplished by sequencing a second library constructed with RNA treated with CIP before the yDcpS treatment, in order to remove the 5′ triphosphate from non–Pol II transcripts. (B) RT-qPCR assay measuring the recovery after streptavidin enrichment of various classes of transcripts such as RNA18S1 rRNA as an example of a processed transcript; ACTB, RPL19, MALAT1, FKBP5, TMSB10, H3C10, and HIST2H3B as examples of capped transcripts; RMRP, RPPH1, and RN7SK as examples of Pol III transcripts (with RN7SK having a 5′ methylated triphosphate and therefore being resistant to CIP treatment; see main text); and ERCC190 and FLUC as examples of spiked-in in vitro transcripts with a defined triphosphorylated 5′ end. The Cq values are available in Supplemental Figure S1F. (C) Example of a Pol II TSS in the TMSB10 locus: The same positions (shaded in pink) are found in the CAGE data set. CIP treatment intensifies the signal, consistent with a Pol II TSS. (D) Example of Pol III TSS corresponding to the start of two vault RNAs (vtRNA1-1 and vtRNA1-2) located on Chr 5. The positions (shaded in pink) are missing in the CAGE data set. CIP treatment reduces the signal, consistent with non–Pol II TSS. In C and D, the tracks correspond to ReCappable-seq, CIP-ReCappable-seq, and CAGE read coverage (number of reads). All libraries were down-sampled to the same number of total mapped reads (63,300,000) to facilitate comparison. The four bottom tracks correspond to read density from public ENCODE DNase-seq from A549 cells (ENCFF473YHH, ENCFF809KIH, ENCFF821UUL, ENCFF961WXW).