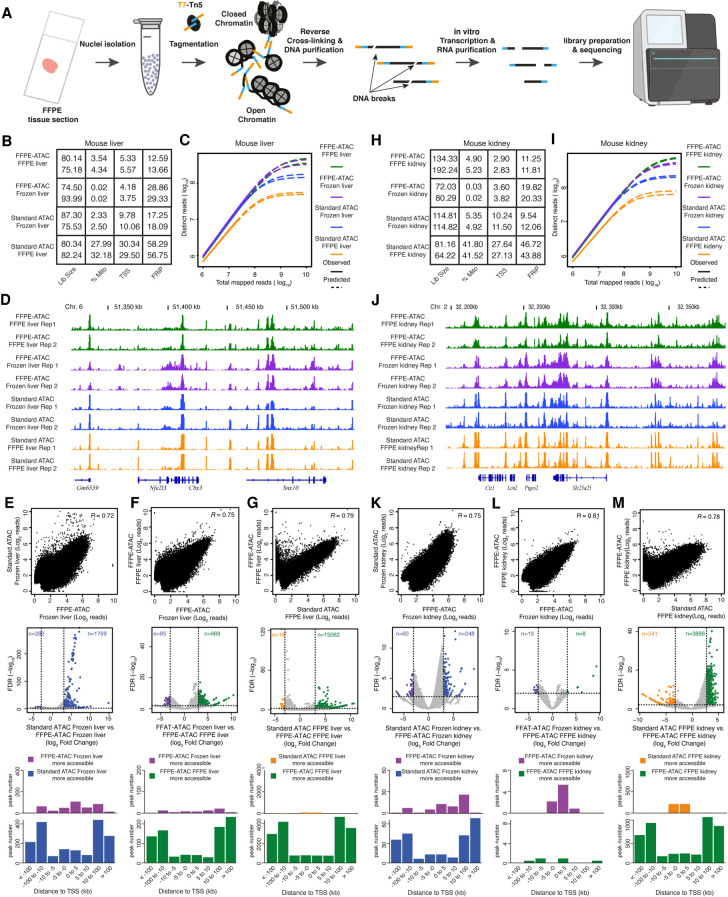
Figure 2.
FFPE-ATAC decodes chromatin accessibility with low cell numbers obtained from FFPE tissue sections. (A) Workflow of FFPE-ATAC. (B) Quality-control metrics of FFPE-ATAC on frozen mouse liver and FFPE mouse liver, and standard ATAC-seq on frozen mouse liver and FFPE mouse liver. (Lib size) Total sequencing reads of sequencing library (million); (%Mito) percentage of mitochondria; (TSS) enrichment score at transcription start sites (TSSs); (FRiP) fraction of reads in peaks. (C,D) Comparison of sequencing library complexity (C) and genome browser tracks (D) from FFPE-ATAC on frozen mouse liver and FFPE mouse liver, and standard ATAC-seq on frozen mouse liver and FFPE mouse liver. (Chr.) Chromosome. (E–G) Comparison of chromatin accessibility from different conditions: standard ATAC-seq on frozen mouse liver versus FFPE-ATAC on frozen mouse liver (E), FFPE-ATAC on frozen mouse liver versus FFPE-ATAC on FFPE mouse liver (F), and FFPE-ATAC on FFPE mouse liver versus standard ATAC-seq on FFPE mouse liver (G). (Top) Genome-wide comparison of accessible chromatin regions. (R) Pearson's correlation. (Middle) Differential peak analysis. (FDR) False-discovery rate. (Bottom) Distribution of the more accessible regions from each condition across TSSs. (H) Quality-control metrics of FFPE-ATAC on frozen mouse kidney and FFPE mouse kidney and standard ATAC-seq on frozen mouse kidney and FFPE mouse kidney. (Lib size) Total sequencing reads of sequencing library (million); (%Mito) percentage of mitochondria; (TSS) enrichment score at TSSs; (FRiP) fraction of reads in peaks. (I,J) Comparison of sequencing library complexity (I) and genome browser tracks (J) from FFPE-ATAC on frozen mouse kidney and FFPE mouse kidney and standard ATAC-seq on frozen mouse kidney and FFPE mouse kidney. (Chr.) Chromosome. (K–M) Comparison of chromatin accessibility from different conditions: standard ATAC-seq on frozen mouse kidney versus FFPE-ATAC on frozen mouse kidney (K), FFPE-ATAC on frozen mouse kidney versus FFPE-ATAC on FFPE mouse kidney (L), and FFPE-ATAC on FFPE mouse kidney versus standard ATAC-seq on FFPE mouse kidney (M). (Top) Genome-wide comparison of accessible chromatin regions. (R) Pearson's correlation. (Middle) Differential peak analysis. (FDR) False-discovery rate. (Bottom) Distribution of the more accessible regions from each condition across TSSs.