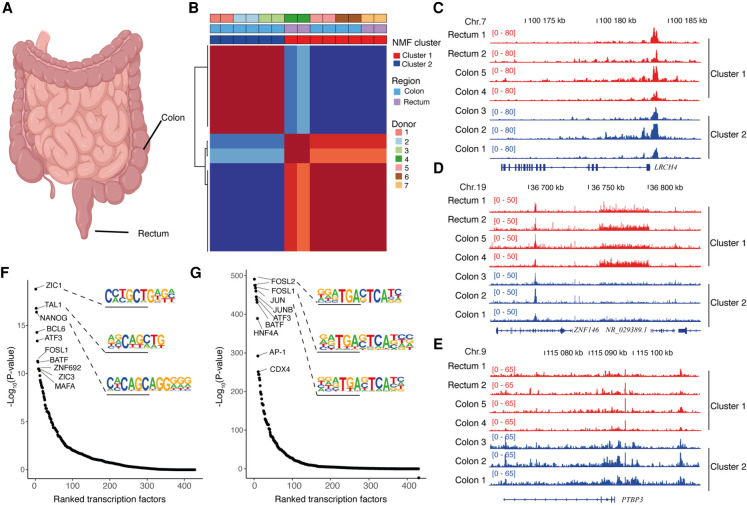
Figure 4.
FFPE-ATAC decodes chromatin accessibility from archived clinical tumor samples. (A) Schematic image (created with BioRender) showing the location of human colorectal cancer (CRC) samples: colon and rectum. (B) Nonnegative matrix factorization (NMF) of chromatin accessibility with FFPE-ATAC from two cases of rectal cancer and five cases of colon cancer, identifying two clusters. (C) Regulatory elements of the CRC marker gene LRCH4 are accessible in both clusters. (D) Representative gene loci that are more accessible in cluster 1, as seen from the differential FFPE-ATAC peaks. (E) Representative gene loci that are more accessible in cluster 2, as seen from the differential FFPE-ATAC peaks. (F) Ranked transcription factors significantly enriched in the specific regulatory elements from cluster 1. (G) Ranked transcription factors significantly enriched in the specific regulatory elements from cluster 2.