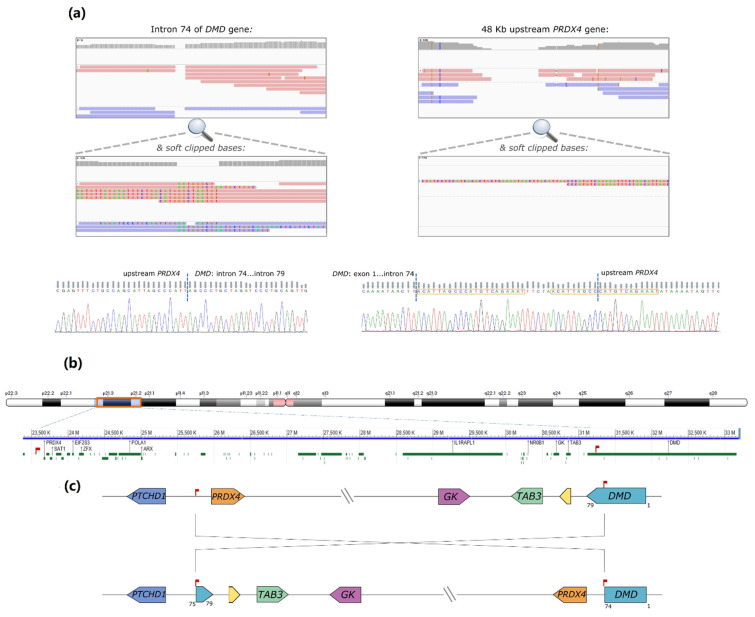
Figure 1.
(a) Low coverage WGS results suggesting a genomic inversion. Visual BAM file inspection showed a possible breakpoint within intron 74 of DMD (1A). Some reads contained soft-clipped bases with homology with a region 48 kb upstream of the PRDX4 gene (Xp22.1). Correspondingly, some partially aligned reads in the PRDX4 region showed homology with inverted intron 74 DMD sequences; (b) Inversion breakpoints were confirmed by Sanger sequencing. In breakpoint 2 (right) the inversion originated a 21 bp duplication (boxed in orange) and the inclusion of a short sequence with no homology to the reference genome. There was also a loss of 9 bp in DMD intron 74 and of 3 bp in the upstream PRDX4 region; (c) Schematic representation of the inversion at the genomic level. The inversion involves Xp22.1 to Xp21.2 (~8 Mb), comprising 74 genes. Breakpoints are localized 48 kb upstream of the PRDX4 gene (breakpoint 1) and within intron 74 of the DMD gene (breakpoint 2).