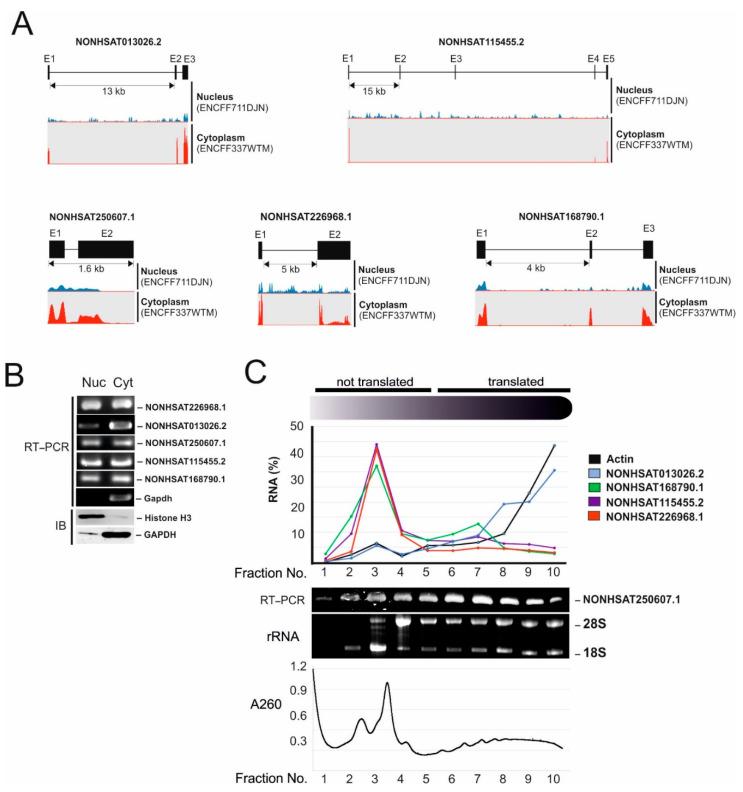
Figure 2.
Identification of micropeptide candidates derived from HCC-specific lincRNAs. (A) Nuclear- (ENCFF711DJN) and cytoplasmic (ENCFF337WTM) RNA-seq of HepG2 cells generated by the ENCODE Consortium were aligned to the reference human genome (GRCh38). SeqMonk was used to quantitate and visualize the data. Peaks in the wiggle plot represent the normalized RNA-seq read coverage on HCC-specific lincRNAs. E: exon. (B) RNA was isolated from the nuclear (Nuc) and cytoplasmic (Cyt) fractions of HepG2 cells and analyzed by RT-PCR. Fractionation quality was measured by immunoblot analysis of THOC5, GAPDH and Histone H3 (Blot). Three independent experiments were performed. (C) HepG2 cytoplasmic lysate was prepared and fractionated on sucrose gradients. The distribution of RNA was calculated using the CT values obtained by qRT-PCR. Isolated RNA was supplied in a gel to determine translated fractions. mRNAs were prepared from the indicated fractions and were applied for Actin, NONHSAT013026.2, NONHSAT168790.1, NONHSAT115455.2, and NONHSAT226968.1 qRT-PCR or NONHSAT250607.1 RT-PCR. A representative absorbance profile at 260 nm was obtained during fractionation of gradients. A replicate is shown in Figure S1.