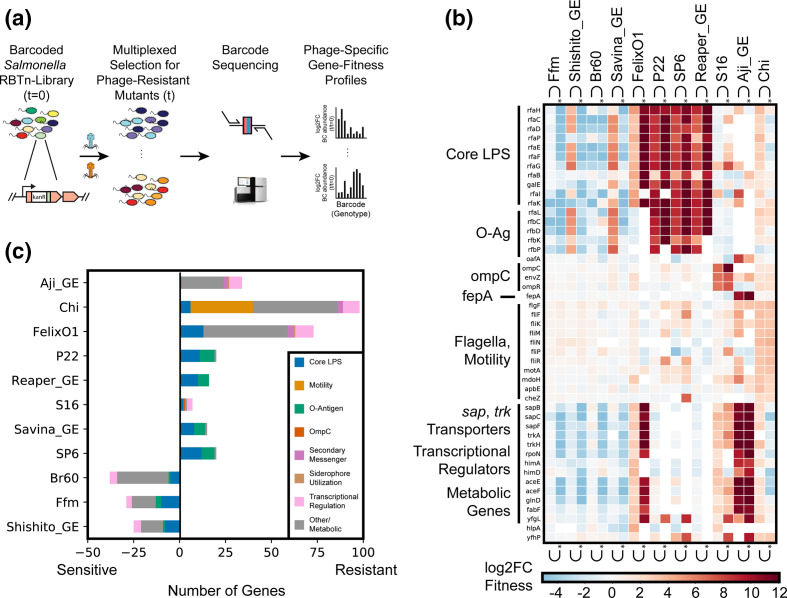
Fig. 1.
Genome-wide screen to identify host factors involved in phage infection. (a) Overview of pooled fitness assays. For additional details, see Methods. Briefly, for each experiment, S. Typhimurium RB-TnSeq library was exposed to a high MOI of one of eleven dsDNA Salmonella phages. Strains were tracked by quantifying the abundance of DNA barcodes associated with each strain by Illumina sequencing. Phage-specific gene fitness profiles were calculated by taking the log2-fold-change of barcode abundances post- (t) to pre- (t=0) phage predation. High fitness scores indicate that loss of genetic function in Salmonella confers fitness against phage predation. (b) Heatmap of top ten high-confidence gene scores per phage are shown (many genes are high-confidence hits to multiple phages). Both planktonic and solid plate data are shown. Three rough-LPS binding phages Br60, Ffm, and Shishito_GE do not infect wild-type MS1868, but can infect specific MS1868 mutants, overall showing negative fitness fitness values in our screen. Noncompetitive, solid agar growth experiments are marked with a (*). (c) Total number of high-scoring genes per phage and their functional role. Input data for Fig. 1(b, c) is found in Dataset S4 and can be recreated using Fig. 1b, c using Supplementary Code.