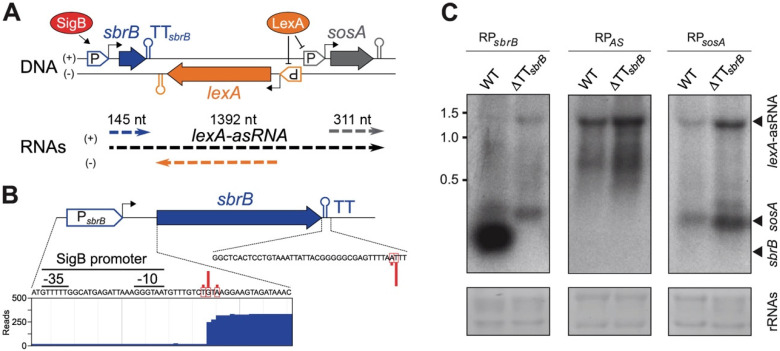
Figure 1.
lexA-asRNA is produced from the SigB-dependent sbrB promoter upon transcriptional read-through of TTsbrB. (A) Schematic representation of the sbrB-lexA-sosA locus according previous transcriptomic maps [4]. ORFs and promoters (P) from sbrB, lexA and sosA genes are represented as blue, orange and gray arrows, respectively. Transcriptional terminators (TT) are illustrated as colored hairpins. Transcripts (RNAs) generated from both DNA strands are represented as dashed arrows. (B) Validation of sbrB mRNA boundaries. The transcriptional start and termination sites of the sbrB were determined by visualizing the S. aureus TSS sequencing data [30] and performing a simultaneous mapping of the 5′ and 3′ mRNA ends by circularization (mRACE) [31]. A Jbrowser image showing the RNA-Seq reads mapping on the sbrB promoter region is also included. The complete transcriptomic map is available at http://rnamaps.unavarra.es, accessed on 8 December 2021 [7]. Red bars represent the frequency of each nucleotide position at the 5′ and 3′ ends identified by mRACE. (C) Northern blots showing the sbrB, lexA-asRNA and sosA mRNA levels expressed from the S. aureus WT and ΔTTsbrB strains. Transcripts were developed using 32P-radiolabelled riboprobes designed to specifically target the sbrB (RPsbrB), lexA-asRNA (RPAS) and sosA (RPsosA) mRNAs. The single-stranded transcript sizes from the RNA Millennium marker are indicated. Midori green-stained ribosomal RNAs are included as loading controls.