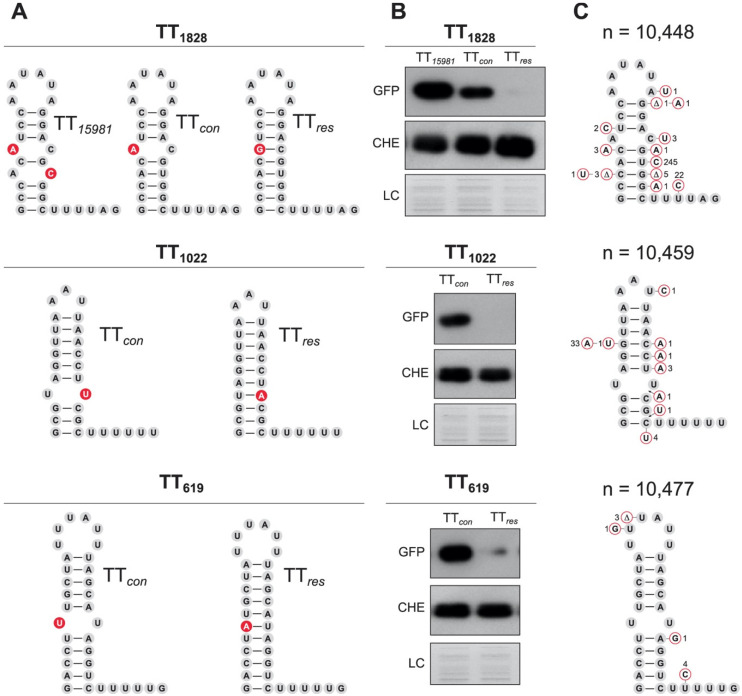
Figure 8.
Evolutionarily selected mispairing nucleotides in TTs are relevant for read-through events. (A) Structures of predicted TTs (1828, 1022 and 619) including mispairings and restored TT variants (res) that compensated the mispairings. Nucleotide changes are colored in red. (B) The TT read-through levels from each variation were monitored by Western blot analyses using the dual fluorescent reporter plasmid shown in Supplementary Figure S6. The GFP expression levels indicate the amount of transcriptional read-through while mCherry monitors the transcript level expressed from the PsbrB promoter. Proteins transferred to nitrocellulose membranes were developed as indicated in Figure 6. Coomassie-stained gel portions are included as loading controls. (C) Identification of natural nucleotide variations in the corresponding TTs. The sequences of the selected TTs were compared by BLASTN against all S. aureus genomes available in the NCBI database. n represents the number of genomes compared for each TT. The nucleotide variations and the number of genomes carrying such variations are indicated in the corresponding TT position.