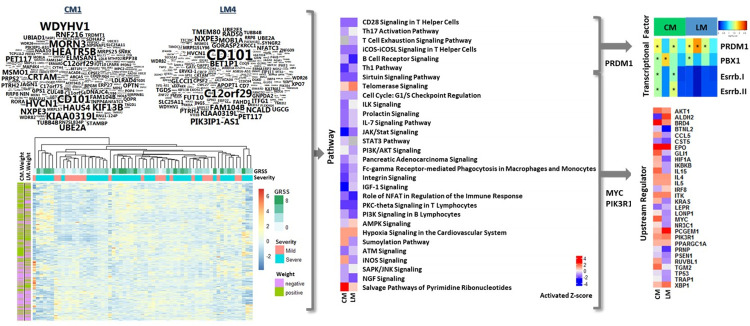
Fig 4. CD4 gene weights, expression, pathways, transcriptional factors and upstream regulators associated with clinical severity in integration models.
Shown are an integration model of CD4, nasal epithelial cells and microbiota (CM) and a model of lymphocytes (LM). Weights generated by integration models are shown in word-clouds. The size of word represents the absolute value of gene weight. Word-clouds of CM & LM consist of genes that have absolute weight is greater than 0.0003. Gene expression are normalized expression levels for the 454 genes selected by univariate analysis; rows represent genes and columns represent samples. Red indicates higher expression, blue indicates low/no expression, green indicates Global RSV Severity Score (GRSS), soft orange indicates mild phenotype, lime green indicates severe phenotype, purple indicates negative weight and olive indicate positive weight. Transcriptional factors associated with severity in CD4 lymphocytes were identified using a hypergeometric test. Four transcriptional factors are shown where p-values were less than 0.05. Ingenuity Pathway Analysis (IPA) was used to identify canonical pathways and upstream regulators represented by genes associated with severity in CD4 lymphocytes. Thirty pathways and upstream regulators are shown where Fisher’s exact test p-values were less than 0.05. Red and blue indicate predicted increased or decreased pathway activation (activation z-score), respectively.