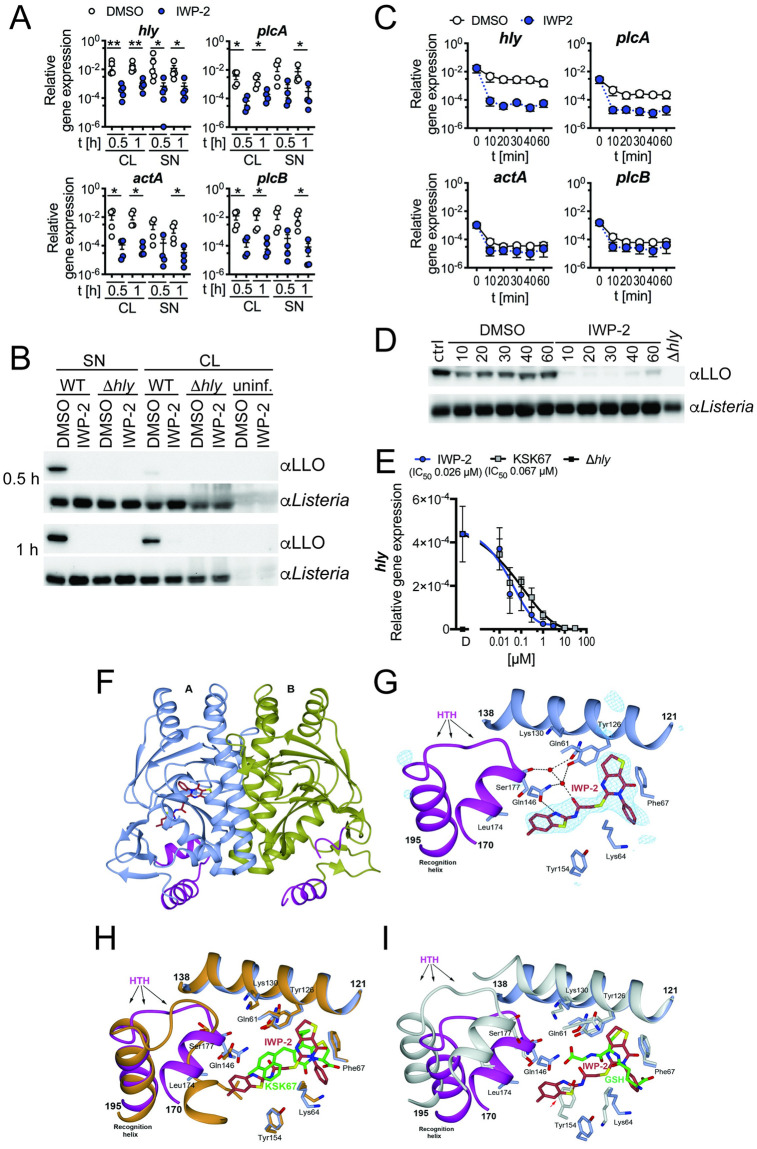
Fig 1. Small molecule occupation of the PrfA coactivator site impairs PrfA-driven virulence gene expression.
(A) Significantly diminished expression of L. monocytogenes virulence genes determined by quantitative RT-PCR in cell lysates (CL) and supernatants (SN) of RAW246.7 macrophages after 0.5 and 1 h of infection in the presence of IWP-2, as compared to DMSO-treated control cells. Data are from 4–5 independent experiments; means +/- sem are indicated. Groups at each condition and time point were compared by Mann-Whitney test. (B) Cell lysates and supernatants of cells infected as in (A) show diminished L. monocytogenes LLO protein expression in the presence of IWP-2 compared to DMSO-treated controls, as determined by western blot. LLO-deficient Δhly L. monocytogenes served as control. Data are representative of 3 independent experiments with similar results. (C) IWP-2 addition rapidly diminished PrfA-controlled L. monocytogenes virulence gene expression in brain heart infusion broth cultures grown at 37°C, as determined by qRT-PCR. Data are means +/- sem of six independent cultures analyzed across three independent experiments. (D) IWP-2 diminished LLO protein expression in L. monocytogenes. Bacteria were grown as in (C) and bacterial lysates analyzed by western blot at the time points indicated [min]. Untreated staring cultures (ctrl), DMSO treatment and LLO-deficient Δhly L. monocytogenes served as controls. Data are representative of 3 independent experiments with similar results. (E) Dose-dependent inhibition of L. monocytogenes hly gene expression in bacterial cultures grown at 37°C, in brain heart infusion broth for 1 h in the presence of IWP-2 or the previously described PrfA inhibitor KSK67(15). DMSO [D] and LLO-deficient Δhly L. monocytogenes served as controls. Data are means +/- sem of 4–6 independent cultures analyzed across 3–4 independent experiments. (F) Crystal structure of PrfA in complex with IWP-2. The PrfA homodimer with IWP-2 bound at the AI site. The monomeric units A and B of PrfA are colored in blue and gold, respectively; the ligand is shown in stick representation. (G) Key local structural features and amino acids in proximity to IWP-2 bound at the AI site. Shown is also the quality of the electron density for IWP-2. The POLDER omit electron density map in blue is calculated 5 Å around each atom of IWP-2 and contoured at 4σ. IWP-2 fits the electron density well and can be modelled into the map with high correlation coefficient [CC(1,3) = 0.85]. (H) Superimposition of the N-terminal domains (residues 2–121) of PrfA structures harboring the 2-pyridone compound KSK67 (PDB ID 6EUT) and IWP-2. Both the N-terminal domain and the C-terminal domain (residues 138–237, which includes the DNA binding HTH motif) superimpose well. The PrfA-2-pyridone structure is shown in orange with KSK67 shown in green stick representation. (I) Superimposition of the N-terminal domains of GSH- (PDB ID 5LRR) and IWP-2-bound PrfA structures. The PrfA:GSH structure is shown in white with GSH shown in green stick representation. Due to the collapsed N-terminal domain in GSH-activated PrfA only parts of the N-terminal domain superimpose. Notably, Tyr154 in GSH-activated PrfA is positioned at the same position as the methyl-benzothiazole group of IWP-2 (red arrow). As a result, in the IWP-2-bound PrfA structure, even though it is folded, the HTH motif has a position not compatible with DNA binding.