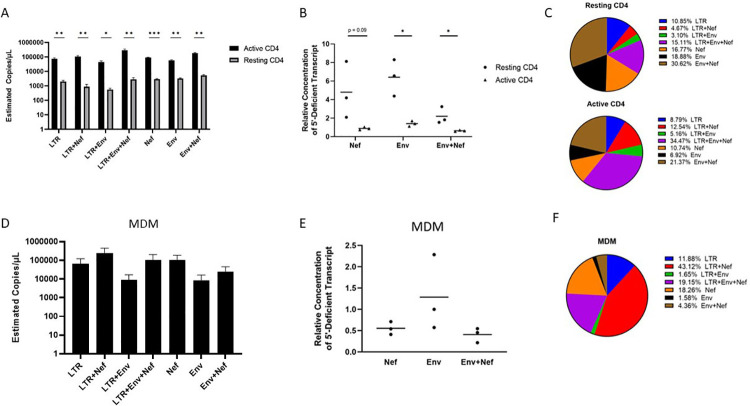
Fig 2. CD4+ T cells and MDMs express aberrant HIV-1 RNAs.
(A) Multiplex RT-ddPCR was performed on RNA prepared from unstimulated and activated CD4+ T cells infected with HIV-1NL4-3 by spinoculation for 90 minutes at an MOI of 0.02 and incubated for 72 hours prior to RNA isolation. HIV-1 infection was limited to a single round by addition of 10 μM saquinavir 30 minutes after spinoculation. Activated CD4+ T cells were cultured with anti-CD3/CD28 beads for 72 hours prior to spinoculation. Estimated copies/μL for transcripts were calculated with the QuantaSoft droplet reader software after manual gating of multiplex RT-ddPCR data (See S3 Fig) and dilution factor correction. (B) Ratios of HIV 5’UTR defective transcripts relative to transcripts with intact 5’UTRs for resting and activated HIV infected CD4+ T cells. (C) Frequencies of detected HIV transcripts as a percentage of all transcripts quantified by multiplex RT-ddPCR for resting (top) and CD3/CD28 activated CD4+ T cells. (D) Multiplex RT-ddPCR performed with RNA from HIV-1NL43-BaL infected MDMs. MDMs were infected with HIV-1NL43-BaL at an MOI of 0.01 for 4 hours by adding virus directly to adherent cell cultures for 48 hours prior to RNA isolation. (E) Ratios of 5’UTR defective transcripts relative to transcripts with intact 5’UTR for HIV infected MDMs. (F) Frequencies of HIV-1 transcripts as a percentage of all transcripts quantified by multiplex RT-ddPCR for HIV infected MDMs. Data represent three independent experiments using cells from three donors. Resting and activated CD4+ T cell sample data are donor matched. Bars represent standard error of the mean. Significance values were generated using multiple unpaired Two-tailed T tests. * denotes p < 0.05; ** denotes p < 0.01, and *** denotes p<0.001.