Abstract
Background
Clubfoot, a congenital deformity that presents as a rigid, inward turning of the foot, affects approximately 1 in 1000 infants and occurs as an isolated birth defect in 80% of patients. Despite its high level of heritability, few causative genes have been identified, and mutations in known genes are only responsible for a small portion of clubfoot heritability.
Questions/purposes
(1) Are any rare gene variants enriched (that is, shared) in unrelated patients with isolated clubfoot? (2) Are there other rare variants in the identified gene (Filamin B) in these patients with clubfoot?
Methods
Whole-exome sequence data were generated from a discovery cohort of 183 unrelated probands with clubfoot and 2492 controls. Variants were filtered with minor allele frequency < 0.02 to identify rare variants as well as small insertions and deletions (indels) resulting in missense variants, nonsense or premature truncation, or in-frame deletions. A candidate deletion was then genotyped in another cohort of 974 unrelated patients with clubfoot (a replication cohort). Other rare variants in the candidate gene were also investigated. A segregation analysis was performed in multigenerational families of individuals with clubfoot to see if the genotypes segregate with phenotypes. Single-variant association analysis was performed using the Fisher two-tailed exact test (exact p values are presented to give an indication of the magnitude of the association).
Results
There were no recurrent variants in the known genes causing clubfoot in this study. A three-base pair in-frame codon deletion of Filamin B (FLNB) (p.E1792del, rs1470699812) was identified in 1.6% (3 of 183) of probands with clubfoot in the discovery cohort compared with 0% of controls (0 of 2492) (odds ratio infinity (inf) [95% CI 5.64 to inf]; p = 3.18 x 10-5) and 0.0016% of gnomAD controls (2 of 125,709) (OR 1.01 x 103 [95% CI 117.42 to 1.64 x 104]; p = 3.13 x 10-8). By screening a replication cohort (n = 974 patients), we found two probands with the identical FLNB deletion. In total, the deletion was identified in 0.43% (5 of 1157) of probands with clubfoot compared with 0% of controls and 0.0016% of gnomAD controls (OR 268.5 [95% CI 43.68 to 2.88 x 103]; p = 1.43 x 10-9). The recurrent FLNB p.E1792del variant segregated with clubfoot, with incomplete penetrance in two families. Affected individuals were more likely to be male and have bilateral clubfoot. Although most patients had isolated clubfoot, features consistent with Larsen syndrome, including upper extremity abnormalities such as elbow and thumb hypermobility and wide, flat thumbs, were noted in affected members of one family. We identified 19 additional rare FLNB missense variants located throughout the gene in patients with clubfoot. One of these missense variants, FLNB p.G2397D, exhibited incomplete penetrance in one family.
Conclusion
A recurrent FLNB E1792 deletion was identified in 0.43% of 1157 isolated patients with clubfoot. Given the absence of any recurrent variants in our discovery phase (n = 183) for any of the known genes causing clubfoot, our findings support that novel and rare missense variants in FLNB in patients with clubfoot, although rare, may be among the most commonly known genetic causes of clubfoot. Patients with FLNB variants often have isolated clubfoot, but they and their family members may be at an increased risk of having additional clinical features consistent with Larsen syndrome.
Clinical Relevance
Identification of FLNB variants may be useful for determining clubfoot recurrence risk and comorbidities.
Introduction
Talipes equinovarus [26], commonly referred to as clubfoot, is one of the most common congenital musculoskeletal birth defects, with a prevalence of one to two affected individuals per 1000 births [34]. Clubfoot presents as a rigid, inward turning of the foot. Approximately 80% of affected individuals have clubfoot as an isolated birth defect, and the remaining 20% have known chromosomal abnormalities or genetic syndromes [9, 18]. Although the etiology of isolated clubfoot is not completely understood, there is strong evidence that genetic factors play a role in abnormal foot development. Pedigree analyses and studies of twins show that clubfoot is heritable, with a recurrence rate among families of approximately 25% and concordance of 0.32 in monozygotic twins compared with only 0.029 in dizygotic twins [6, 12, 22, 28].
Candidate gene association studies, genome-wide association studies, copy number analysis, and exome sequencing have identified the genetic causes of isolated clubfoot. These studies have recognized genes associated with clubfoot including those involved in limb bud initiation (Hox genes) [5, 11, 29], hindlimb formation (the PITX1-TBX4 pathway) [1, 2, 4], and muscle contraction (TPM1, TPM2, and TNNC2) [30, 32]. However, despite the identification of genes associated with clubfoot, gene mutations explain only a very small percentage of clubfoot heritability.
In this study, we therefore asked: (1) Are any rare gene variants enriched (that is, shared) in unrelated patients with isolated clubfoot? (2) Are there other rare variants in the identified gene, Filamin B (FLNB), in these patients with clubfoot?
Materials and Methods
Overview of Study Design
To answer our study questions, we performed a whole exome sequencing analysis to identify new genetic causes of clubfoot in unrelated patients with isolated clubfoot. In this study, 1157 unrelated Caucasian patients with idiopathic clubfoot were included. Among these, 183 patients were included in the discovery cohort for initial whole exome sequencing and the remaining 974 patients were either genotyped or sequenced as the replication cohort. A cohort with 2492 Caucasian patients with unrelated phenotypes was used as the control group. To address our first question regarding whether any rare gene variants enriched in unrelated patients with isolated clubfoot, we analyzed variants that are nonsense, missense, splice-site, insertion, or deletion with a gnomAD minor allele frequency < 0.02 and only present in three or more individuals (enriched) with clubfoot (n = 183) and absent in the controls (n = 2492). The variant of interest was then screened in the replication cohort (n = 974). Segregation analysis of the variant of interest was performed if the additional family members were available. To assess whether there were other rare variants in the identified gene in these patients with clubfoot, we analyzed variants in the identified gene (FLNB) with a gnomAD minor allele frequency < 0.02 and absent in the controls (n = 2492). Segregation analysis of the variant of interest was performed if the additional family members were available.
Patient Samples
We recruited 1157 unrelated Caucasian patients with idiopathic clubfoot at St. Louis Children’s Hospital in St. Louis, MO, USA, and Shriners Hospital in Houston, TX, USA. The composition of the patients contains an approximately 2:1 male to female ratio. The patients were younger than 18 years old. We included all patients evaluated for idiopathic clubfoot by an orthopaedic surgeon (MBD), but these patients did not have an underlying known genetic diagnosis. For this study, the clubfoot diagnosis required rigid hindfoot equinus, hindfoot varus, midfoot supination, and midfoot cavus deformities. Patients with additional birth defects, known genetic syndromes, or a parent-reported developmental delay were excluded. Blood or saliva samples were obtained from each patient, and DNA was isolated using DNA Genotek kits (blood: prepIT-L2P PT-L2P; saliva: Oragene-Discover OGR-500 and ORAcollect for Pediatrics OC-175). DNA quality was assessed by 260/280 values and by visualizing genomic DNA via agarose gel electrophoresis. In all, 183 patients were included in the discovery cohort for initial analysis of whole exome sequencing, and the other 974 patients were included as the replication cohort. The discovery cohort consisted of patients who were recruited between 2005 and 2012, and the replication cohort was recruited between 2012 and 2020. Both cohorts had a 2:1 male to female ratio.
Control Samples
The controls consisted of unrelated Caucasian individuals with Alzheimer disease, amyotrophic lateral sclerosis, and adolescent idiopathic scoliosis of equal male and female representation, and aged 12 to 65 years old.
Whole Exome Sequencing and Analysis
Whole exome sequencing uses next-generation sequencing technology to sequence the exonic regions of the human genome. It was performed on unrelated Caucasian patients with idiopathic clubfoot and unrelated control Caucasian patients. Raw sequencing reads were aligned to the human genome reference (GRCh37) using Burrows-Wheeler Aligner (BWA-MEM) version 0.7.15 [23]. Binary Alignment Map (BAM) files were sorted, and polymerase chain reaction duplicate reads were marked using Picard MarkDuplicates (version 2.9.0). GATK version 3.5 was used for the following data processing. Small insertions and deletions (indels) were realigned using GATK RealignerTargetCreator and IndelRealigner using a known indel variant-sites database (Mills and 1kg indels from the GATK resource bundle [14]). The base quality score was then recalibrated using GATK BaseRecalibrator and PrintReads to generate a final high-quality BAM file. Variant calling of single nucleotide variants and indels were finally generated, first for each single sample and then combining all samples using the joint genotyping method, described in GATK Best Practices [15]. Variant calls were then recalibrated and filtered using the GATK Variant Quality Score Recalibration method. A final Variant Call Format file with high-quality variants was then annotated using the Gencode version 19 database [13]. The analysis was restricted to rare, coding sequence-altering variants. For this study, we defined this as nonsense, missense, splice-site, insertion, or deletion variants with a gnomAD minor allele frequency < 0.02. For the initial variant enrichment discovery (shared in multiple individuals), we further winnowed the variants with the following condition: present in three or more individuals with clubfoot (n = 183) and absent in the controls (n = 2492).
Combined Annotation–Dependent Depletion (CADD) [20] was used to evaluate the predicted variant pathogenicity. The CADD scaled score represents the ranking of the predicted pathogenicity in order of magnitude, with a CADD scaled score 20 as top 1% deleterious variants and CADD scaled score 30 as top 0.1% deleterious variants in the genome [20].
Polymerase Chain Reaction Screening of the Replication Cohort
We used a separate cohort of unrelated patients with clubfoot (n = 974) to test whether the finding in the discovery cohort (n = 183) could be replicated. Two sets of allele-specific primers were designed to screen this replication cohort for the in-frame deletion in Filamin B (FLNB): one annealing to the wild-type sequence producing a 456-bp amplicon and the other annealing to sequences producing a 308-bp amplicon. Polymerase chain reaction (PCR) products were examined by agarose gel electrophoresis. Samples indicating a deletion were validated through Sanger sequencing.
Ethical Approval
The study protocol was approved by the institutional review board of Washington University in St. Louis, MO, USA (number 201102118). All patients and/or parents gave informed consent.
Statistical Analysis
The single-variant association analysis was performed using the Fisher two-tailed exact test. Exact p values are presented to give an indication of the magnitude of the association.
Results
A Recurrent, Rare Indel in FLNB Is Identified in Unrelated Patients with Idiopathic Clubfoot
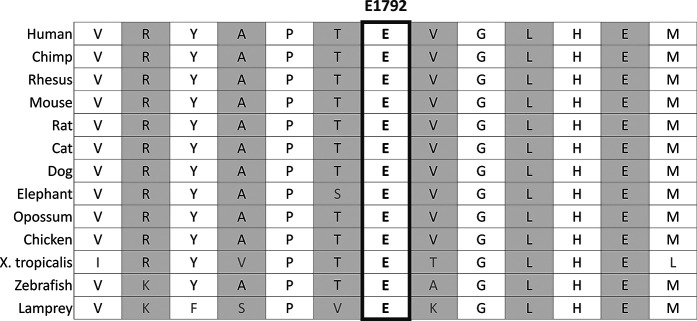
From our discovery (n = 183) and control cohorts (n = 2492), we identified 13 rare nonsynonymous or indel variants presenting in three or more individuals with clubfoot and absent in the controls. The variant with the lowest minor allele frequency in gnomAD (8.1 x 10-6) was a three-base pair, in-frame codon deletion within the repeat domain of FLNB (p.E1792del; NM_001457.4:c.5375_5377del) (Fig. 1). This heterozygous variant resulted in the deletion of a highly conserved glutamine residue (Fig. 2) that was validated by Sanger sequencing in all three probands. It was present in 1.6% (3 of 183) of probands with clubfoot compared with 0% of controls (0 of 2492) (odds ratio infinity (inf) [95% CI 5.64 to inf]; p = 3.18 x 10-5) and 0.0016% (2 of 125,709) of gnomAD controls (OR 1.01 x 103 [95% CI 117.42 to 1.64 x 104]; p = 3.13 x 10-8). In the replication cohort of 974 unrelated patients with clubfoot, two individuals were found to have the exact same codon deletion. When we combined the replication cohort (n = 974) with the discovery cohort (n = 183), the deletion was identified in 0.43% (5 of 1157) of probands with clubfoot compared with 0.0016% of controls (gnomAD) (OR 268.5 [95% CI 43.68 to 2.88 x 103]; p = 1.43 x 10-9).
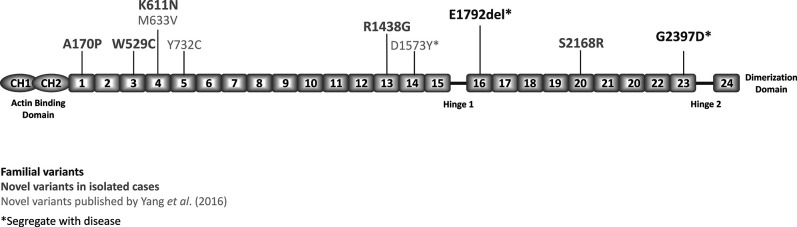
Fig. 1.
This figure displays variants in FLNB in patients with clubfoot. Domains where Larsen syndrome mutations have been reported are highlighted in blue. A color image accompanies the online version of this article.
Fig. 2.
Human FLNB E1792 is conserved across species.
Sanger sequencing revealed that the FLNB codon deletion segregated with clubfoot, with incomplete penetrance in three families (Fig. 3A-C). This FLNB codon deletion was found in nine individuals (two female; seven male) from five families. Clinically, 7 of 8 patients with genotype-confirmed clubfoot had bilateral clubfoot and one had unilateral clubfoot (Table 1). Upper extremity abnormalities, including hypermobile elbows and thumbs, as well as wide and flat thumbs, were reported in affected members of one family but were not present in individuals from any other family. One family member reported mild scoliosis. Our results show that patients carrying FLNB variants often have isolated clubfoot, but they and their family members may be at an increased risk of having additional clinical features consistent with Larsen syndrome (Table 1).
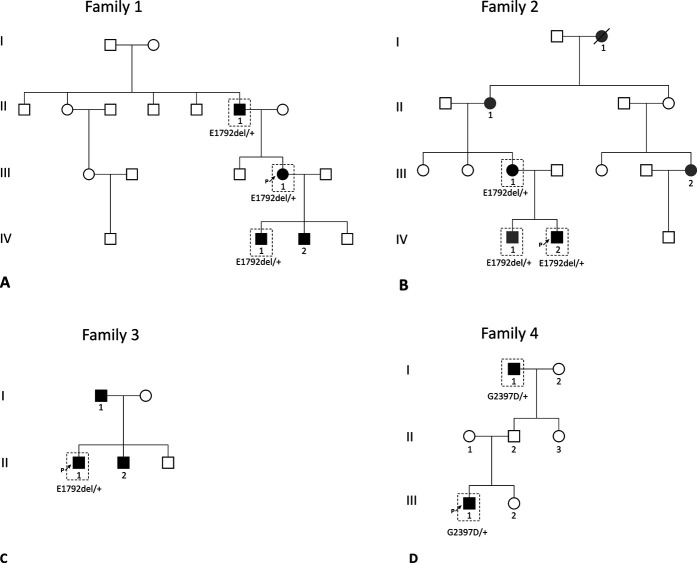
Fig. 3.
A-D This figure shows the pedigrees of families with FLNB variants. Black indicates talipes equinovarus, gray indicates a related phenotype, and the dashed box indicates genotyped individuals. (A) Depicts Family 1 with E1792del, (B) depicts Family 2 with E1792del, (C) depicts Family 3 with E1792del, and (D) depicts Family 4 with the novel G2397D variant.
Table 1.
Clinical features of families with FLNB variants
| Family | Individual | Variant | Clubfoot phenotype | Other phenotypes/comorbidities |
| 1 | II-1 | E1792del | Bilateral clubfoot | |
| III-1 (proband) | E1792del | Bilateral clubfoot | ||
| IV-1 | E1792del | Bilateral clubfoot | ||
| IV-2 | Not genotyped | Bilateral clubfoot | ||
| 2 | I-1 | Not genotyped | None | Toe syndactyly |
| II-1 | Not genotyped | None | Thumb hypermobility | |
| III-1 | E1792del | Bilateral clubfoot | Mild scoliosis and flat thumbs with wide nailbeds | |
| III-2 | Not genotyped | None | Toe syndactyly | |
| IV-1 | E1792del | None | Curly or overlapping toes, flat feet, and thumb hypermobility | |
| IV-2 (proband) | E1792del | Bilateral clubfoot | ||
| 3 | I-1 | Not genotyped | Bilateral clubfoot | |
| II-1 (proband) | E1792del | Left clubfoot | ||
| II-2 | Not genotyped | Right clubfoot | ||
| 4 | I-1 | G2397D | Bilateral clubfoot | |
| III-1 (proband) | G2397D | Bilateral clubfoot | Elbow hypermobility |
Identification of Additional Novel and Rare FLNB Variants in Clubfoot
To determine whether there are other rare variants in FLNB in patients with clubfoot, a single novel missense variant, G2397D, was identified and was absent from the gnomAD database (Fig. 3D). FLNB G2397D segregated with clubfoot in an affected grandfather and had a CADD scaled score of 33 [20], which predicted this variant as the top 0.1% of the most deleterious variants in the genome.
In the replication cohort, we further identified five additional novel missense variants that were not present in gnomAD or dbSNP (Table 2). We also identified 13 rare missense variants that were present in low frequency in gnomAD or dbSNP while not present in our control group (Table 3). None were reported as pathogenic in ClinVar [25], although four were listed as variants of uncertain significance, and two were listed as likely benign.
Table 2.
Novel FLNB missense variants in isolated patients with clubfoot
| Variant | Position | Base change | ID | gnomAD MAF | Protein domain | CADD phred |
| A170P | chr3:58062988 | G>C | Novel | Novel | Repeat 1 | 28.1 |
| W529C | chr3:58089789 | G>C | Novel | Novel | Repeat 3 | 25.0 |
| K611N | chr3:58092492 | G>C | Novel | Novel | Repeat 4 | 24.0 |
| R1438G | chr3:58116557 | C>G | Novel | Novel | Repeat 13 | 20.6 |
| S2168R | chr3:58139238 | C>G | Novel | Novel | Repeat 20 | 24.8 |
| G2397D | chr3:58149049 | G>A | Novel | Novel | Repeat 23 | 33 |
ID = dbSNP identification number; CADD phred = Combined Annotation Dependent Depletion phred scaled score, predicting the deleteriousness of variants.
Table 3.
Rare FLNB missense variants identified in isolated patients with clubfoot
| Variant | Position | Base change | ID | gnomAD MAF | Protein domain | CADD phred | ClinVar |
| R89G | chr3:57994556 | C>G | rs755518501 | 3.98E-06 | CH1 | 23.0 | |
| I92V | chr3:57994565 | A>G | rs62622011 | 7.61E-04 | CH1 | 23.2 | Likely benign |
| I430M | chr3:58084580 | C>G | rs147854989 | 1.38E-04 | Repeat 2 | 25.0 | VUS |
| D435N | chr3:58084593 | G>A | rs781162510 | 6.38E-05 | Repeat 2 | 20.8 | |
| T669A | chr3:58094248 | A>G | rs147481678 | 7.07E-05 | Repeat 5 | 14.0 | VUS |
| V1195M | chr3:58109276 | G>A | rs200993986 | 6.51E-04 | Repeat 10 | 23.1 | Likely benign |
| D1396N | chr3:58112453 | G>A | rs766688604 | 2.83E-05 | Repeat 12 | 27.3 | |
| R1438C | chr3:58116557 | C>T | rs1376465311 | 2.49E-05 | Repeat 13 | 23.4 | |
| E1632K | chr3:58124041 | G>A | rs768316224 | 1.77E-05 | Repeat 15 | 27.3 | |
| R2010C | chr3:58134516 | C>T | rs138034708 | 1.98E-04 | Repeat 19 | 33.0 | |
| L2052V | chr3:58135639 | C>G | rs372726521 | in dbSNP | Repeat 19 | 13.1 | VUS |
| T2166M | chr3:58139231 | C>T | rs199939739 | 5.66E-05 | Repeat 20 | 33.0 | VUS |
| R2346C | chr3:58148895 | C>T | rs149638325 | 6.01E-05 | Repeat 22 | 35.0 |
ID = dbSNP identification number; MAF = minor allele frequency; CADD phred = Combined Annotation Dependent Depletion phred scaled score, predicting the deleteriousness of variants; ClinVar = a database with the relationships between human variants and phenotypes; VUS = variant of unknown significance.
Discussion
Talipes equinovarus, or clubfoot, is one of the most common congenital musculoskeletal birth defects [34]. Previous pedigree analyses and studies of twins showed that clubfoot is heritable [6, 12, 22, 28], and genes involved in limb bud initiation (Hox genes) [5, 11, 29], hindlimb formation (the PITX1-TBX4 pathway) [1 , 2, 4], and muscle contraction (TPM1, TPM2, and TNNC2) [30, 32] have been identified as being associated with clubfoot through different approaches. However, mutations in these genes are present in only a small percentage of patients with clubfoot. We identified a recurrent FLNB in-frame codon deletion in unrelated patients with isolated clubfoot, including whole-exome data from a discovery cohort of 183 patients and PCR genotyping data from a replication cohort with 974 patients. This FLNB p.E1792del was also present in other affected family members with clubfoot (that is, segregated with clubfoot) in two families, with incomplete penetrance (that is, some family members without clubfoot also carry this variant). Multiple novel and rare FLNB variants were identified in patients with or without a family history of clubfoot. These results suggest that identifying variants in FLNB in multigenerational families with members who have clubfoot may be important to determine clubfoot recurrence risk and to predict comorbidities.
Limitations
This study has several limitations. First, whole exome sequencing only covers the coding regions, and variants in the promoter or intronic region are not included. There may be other clubfoot-related variants outside the exon regions that could be identified through whole-genome sequencing in the future. Second, the sample size was relatively small (n = 183) in our discovery cohort in the whole exome sequencing analysis. This may at least partially explain why we did not find an overall enrichment of FLNB missense variants in patients with clubfoot compared with controls. In addition, many patients carrying these missense variants did not have additional family members to test for segregation and pathogenicity. Third, the mechanism of how these FLNB variants contribute to the pathogenesis of clubfoot is not fully understood, although there are some potential mechanisms (which we discuss later in the Discussion). These will require additional in vivo and in vitro studies. Fourth, the oligogenic contribution of variants was not analyzed in this study, but a large collection of samples will be required to address this. Fifth, sex was not considered in our study because the sample size was too small to be further split by sex. Given that this is a dicovery study, a later study can investigate the specific role of sex.
A Recurrent, Rare Indel in FLNB Is Identified in Unrelated Patients with Idiopathic Clubfoot
In this study, we reported a recurrent FLNB in-frame deletion in 0.43% of all unrelated patients with isolated clubfoot. The absence of any recurrent variants in our discovery phase (n = 183) for any of the known genes causing clubfoot compared with FLNB as well as our previously published incidence data [1, 2, 5] suggest this recurrent deletion may represent as one of the most common genetic factors for clubfoot. The frequency of this deletion is likely even higher in families with clubfoot because the frequency was 1.6% in our exome sequence discovery cohort, which was enriched in patients (presented in multiple individuals) with a family history of clubfoot. Although this recurrent FLNB p.E1792del variant has not been reported to cause clubfoot, three FLNB variants have been reported in patients with clubfoot [38]. Yang et al. [38] described a large Chinese family with isolated clubfoot with a missense variant in Repeat 14 of FLNB that segregated with clubfoot in six affected individuals. The authors also sequenced the data of 53 patients with sporadic clubfoot and found two heterozygous FLNB missense variants in Repeats 4 and 5. None had phenotypic abnormalities other than clubfoot [38]. However, unlike our FLNB p.E1792del variant, none of the FLNB variants reported by Yang et al. [38] were recurrent.
FLNB has been implicated in several types of human skeletal dysplasia, which further supports its role in clubfoot. The human Filamin family contains three homologous actin-binding cytoplasmic proteins encoded by Filamin A (FLNA), Filamin B (FLNB), and Filamin C (FLNC). Filamin proteins crosslink actin fibrils and link the extracellular matrix, cell membrane, and cytoskeleton [24, 27, 39]. Filamins interact with numerous cytoplasmic and transmembrane proteins and play a major role in mechanotransduction in the cytoplasm. FLNA and FLNB are expressed ubiquitously throughout many tissues, and FLNC is primarily expressed in striated and cardiac muscle. Mutations in FLNA and FLNC lead to disorders involving the central nervous, circulatory, and skeletal systems [39]. However, to date, mutations in FLNB have only been associated with skeletal disorders [36]. Similar to all filamins, the FLNB protein contains an N-terminal F-actin binding domain and rod segment comprising 24 immunoglobulin-like repeat domains with two flexible hinge regions between Repeats 15 and 16 and between Repeats 23 and 24 [17]. These hinge regions confer flexibility on filamin dimers. The C-terminal domain (Repeat 24) facilitates dimerization. Functional FLNB protein is present as either a homodimer or heterodimer.
Variants in FLNB cause several Mendelian-inherited skeletal dysplasias, including boomerang dysplasia, atelosteogenesis Type I and III, spondylocarpotarsal synostosis, and Larsen syndrome, and have been implicated in scoliosis [19, 21, 36]. Clubfoot is often an associated phenotype in these syndromes. However, in these disorders, pathogenic FLNB mutations are primarily found in the CH2 domain and in domains near hinge-1 [10]. There are clear genotype-phenotype correlations, with mutations that cause specific disorders often clustering in the same domains. Boomerang dysplasia, characterized by dwarfism and bowed limbs, is lethal in the neonatal period and is caused by heterozygous missense mutations located exclusively in the CH2 domain of FLNB [8, 36]. Atelosteogenesis Type I and III are perinatally lethal skeletal dysplasias characterized by severe short-limbed dwarfism and are caused by mutations in the CH2 domain of FLNB, as well as mutations in Repeats 14 and 15. Biallelic truncations of FLNB lead to spondylocarpotarsal synostosis, which is an autosomal recessive disorder characterized by fused vertebrae and carpal and tarsal joints [21, 37]. Larsen syndrome is less severe than the discussed skeletal dysplasias and is characterized by large-joint dislocations, supernumerary carpal and tarsal bones, and craniofacial abnormalities such as wide-spaced eyes and a prominent forehead [16, 35]. Larsen syndrome is caused by missense mutations and in-frame codon deletions; however, most causative mutations are found in the CH2 domain and near the hinge-1 region in Repeats 14, 15, and 17. Clubfoot is a common, but not essential, feature of Larsen syndrome.
The FLNB p.E1792del variant identified in our patients with clubfoot resides near the hinge-1 region in Repeat 16 (Fig. 1). Nearby pathogenic mutations in Repeats 13, 14, 15, 17, and 23 are known to cause Larsen syndrome [16, 36] (Fig. 1). This residue is highly conserved across different species, suggesting that it is an important residue (Fig. 2), and deleting this residue in FLNB is likely to be deleterious for the protein function. We suggest that the in-frame codon deletion observed in our patients with clubfoot results in a less severe phenotype than the nearby missense mutations that cause Larsen syndrome. Although patients with Larsen syndrome often have clubfoot as a phenotype, almost all of our patients and their family members had no additional phenotypes to support a diagnosis of Larsen syndrome. However, members of one of our families had the variable upper extremity abnormalities and wide thumbs that are characteristic of Larsen syndrome. Of note, the identical FLNB protein p.E1792del variant was recently reported in ClinVar as likely pathogenic by a single submitter related to its segregation in multiple affected family members with autosomal dominant inheritance of knee dislocation, patellar hypoplasia, and limited knee flexion and extension [25]. Therefore, the incomplete penetrance and variable expressivity of the FLNB p.E1792del variant results in a phenotypic spectrum spanning from isolated clubfoot to the complete Larsen syndrome phenotype. The high frequency of the FLNB p.E1792del variant in our cohort suggests that isolated clubfoot may be the most commonly expressed phenotype.
Identification of Additional Novel and Rare FLNB Variants in Clubfoot
Given the finding of a recurrent FLNB deletion in patients with clubfoot, we further investigated whether other rare variants in FLNB are present in patients with clubfoot. In the same dataset, we identified one novel missense variant in FLNB that segregated in a small family, and rare or novel missense variants in 18 patients with sporadic clubfoot that were not present in the control group (five novel; 13 rare). Although there was no overall enrichment of FLNB missense variants in patients with clubfoot compared with controls, this may be related to the small sample size. We suspect that some of the rare FLNB missense variants we identified are pathogenic, as suggested by the segregation of FLNB p.G2397D with clubfoot in three generations and its absence in gnomAD. FLNB p.G2397D is located in Domain 23, which also contains pathogenic variants of Larsen syndrome. The other novel FLNB missense variants we discovered in our patients, which were also absent from gnomAD, are located in Domains 1, 3, 4, 13, and 20. Many of these patients did not have additional family members to test for segregation; therefore, we cannot confirm the pathogenicity of these variants. The occurrence in apparently nonfamilial individuals with clubfoot suggests that these alleles may confer an incompletely penetrant risk of clubfoot. When we combine the two variants reported by Yang et al. [38] with those described here, seven novel FLNB missense variants have been reported in patients with sporadic clubfoot.
There are several proposed mechanisms by which FLNB variants cause abnormal skeletal development. In vitro and in vivo studies demonstrated that FLNB deficiency results in delayed ossification in the long-bone growth plate, ossification of the intervertebral discs, dysregulation of chondrocyte development, and hypomobility of osteoblasts [36]. Interestingly, FLNB has also been implicated in muscle differentiation [7, 31]. As discussed, the mechanisms underlying clubfoot are not fully understood; however, there is evidence that altered muscle development may play a role. Variations in sarcomeric genes such as MYH3, MYH8, TNNI2, TNNI3, and TPM2 are known to cause some types of distal arthrogryposis [3, 30], which have clubfoot as a common feature. Moreover, studies have shown that variants in muscle sarcomeric genes increase the risk of isolated clubfoot [32, 33].
Conclusion
We identified a recurrent FLNB in-frame codon deletion that accounts for 0.43% of unrelated patients with isolated clubfoot from a collection of 1157 unrelated patients. Given the absence of any recurrent variants in our discovery phase (n = 183) for any of the known genes causing clubfoot, this recurrent deletion may represent one of the most common genetic causes of clubfoot. Three of these occurred in patients with a family history of clubfoot who had autosomal dominant inheritance with reduced penetrance, and rarely, variable expressivity with other features of Larsen syndrome, such as wide thumbs and joint hypermobility. We also found a novel FLNB variant segregating with clubfoot and multiple novel and rare missense variants in patients with sporadic clubfoot. Combining these results with published data, there are now three reported FLNB variants that segregate with clubfoot: p.E1792del (two families and three isolated patients), G2397D (one family), and D1573Y (one family, from the study by Yang et al. [38]). Currently, there is insufficient evidence to determine the mechanism by which FLNB pathogenic variants may contribute to clubfoot; therefore, more research is needed. Altogether, there is compelling evidence supporting an important role for FLNB in the pathogenesis of familial and isolated clubfoot. Identifying FLNB variants is important to determine the risk of recurrent clubfoot and may be useful for predicting associated comorbidities.
Acknowledgment
We thank the patients and their family members who participated in this study.
Footnotes
Two authors (MBD, CAG) were supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases under Award Number R01AR067715. Research reported in this publication was supported Eunice Kennedy Shriver National Institutes of Child Health and Human Development of the National Institutes of Health under Award Number P01HD084387, Washington University Institute of Clinical and Translational Sciences grant UL1 TR002345 from the National Center for Advancing Translational Sciences of the National Institutes of Health, and the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health under Award Number P50HD103525 to the Intellectual and Developmental Disabilities Research Center at Washington University. One author (WC) was supported by the National Institute of Mental Health of the National Institutes of Health (T32-MH014677). One author (JTH) was funded by R01HD043342. Computations were performed using the facilities of the Washington University Center for High Performance Computing, which were partially funded by NIH grants 1S10RR022984-01A1 and 1S10OD018091-01.
Each author certifies that there are no commercial associations (consultancies, stock ownership, equity interest, patent/licensing arrangements, etc.) that might pose a conflict of interest in connection with the submitted article.
All ICMJE Conflict of Interest Forms for authors and Clinical Orthopaedics and Related Research® editors and board members are on file with the publication and can be viewed on request.
Ethical approval for this study was obtained from Washing University in Saint Louis, St. Louis, MO, USA (number 201102118).
This work was performed at Washington University, St. Louis, MO, USA.
Contributor Information
Ashley Quiggle, Email: ashley.quiggle@gmail.com.
Wu-Lin Charng, Email: charngw@wustl.edu.
Lilian Antunes, Email: antunes@wustl.edu.
Momchil Nikolov, Email: m.nikolov@wustl.edu.
Xavier Bledsoe, Email: Xavier.bledsoe@vanderbilt.edu.
Jacqueline T. Hecht, Email: Jacqueline.T.Hecht@uth.tmc.edu.
Matthew B. Dobbs, Email: mdobbs@paleyinstitute.org.
References
- 1.Alvarado DM, Aferol H, McCall K, et al. Familial isolated clubfoot is associated with recurrent chromosome 17q23.1q23.2 microduplications containing TBX4. Am J Hum Genet. 2010;87:154-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarado DM, Buchan JG, Frick SL, Herzenberg JE, Dobbs MB, Gurnett CA. Copy number analysis of 413 isolated talipes equinovarus patients suggests role for transcriptional regulators of early limb development. Eur J Hum Genet. 2013;21:373-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alvarado DM, Buchan JG, Gurnett CA, Dobbs MB. Exome sequencing identifies an MYH3 mutation in a family with distal arthrogryposis type 1. J Bone Joint Surg Am. 2011;93:1045-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alvarado DM, McCall K, Aferol H, et al. Pitx1 haploinsufficiency causes clubfoot in humans and a clubfoot-like phenotype in mice. Hum Mol Genet. 2011;20:3943-3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alvarado DM, McCall K, Hecht JT, Dobbs MB, Gurnett CA. Deletions of 5' HOXC genes are associated with lower extremity malformations, including clubfoot and vertical talus. J Med Genet. 2016;53:250-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barker S, Chesney D, Miedzybrodzka Z, Maffulli N. Genetics and epidemiology of idiopathic congenital talipes equinovarus. J Pediatr Orthop. 2003;23:265-272. [PubMed] [Google Scholar]
- 7.Bello NF, Lamsoul I, Heuze ML, et al. The E3 ubiquitin ligase specificity subunit ASB2beta is a novel regulator of muscle differentiation that targets filamin B to proteasomal degradation. Cell Death Differ. 2009;16:921-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bicknell LS, Morgan T, Bonafe L, et al. Mutations in FLNB cause boomerang dysplasia. J Med Genet. 2005;42:e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brewer C, Holloway S, Zawalnyski P, Schinzel A, FitzPatrick D. A chromosomal deletion map of human malformations. Am J Hum Genet. 1998;63:1153-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniel PB, Morgan T, Alanay Y, et al. Disease-associated mutations in the actin-binding domain of filamin B cause cytoplasmic focal accumulations correlating with disease severity. Hum Mutat. 2012;33:665-673. [DOI] [PubMed] [Google Scholar]
- 11.Dobbs MB, Gurnett CA, Pierce B, et al. HOXD10 M319K mutation in a family with isolated congenital vertical talus. J Orthop Res. 2006;24:448-453. [DOI] [PubMed] [Google Scholar]
- 12.Engell V, Nielsen J, Damborg F, et al. Heritability of clubfoot: a twin study. J Child Orthop. 2014;8:37-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gencode version 19. Available at: https://www.gencodegenes.org/human/release_19.html. Accessed July 5, 2017.
- 14.Genome Analysis Toolkit (GATK) resource bundle. Available at: https://console.cloud.google.com/storage/browser/gatk-legacy-bundles/b37?pageState=(%22StorageObjectListTable%22:(%22f%22:%22%255B%255D%22))&prefix=&forceOnObjectsSortingFiltering=false. Accessed August 4, 2021.
- 15.Genome Analysis Toolkit (GATK v3.5) best practices. Available at: https://gatk.broadinstitute.org/hc/en-us/sections/360007226651-Best-Practices-Workflows. Accessed July 1, 2017.
- 16.Girisha KM, Bidchol AM, Graul-Neumann L, et al. Phenotype and genotype in patients with Larsen syndrome: clinical homogeneity and allelic heterogeneity in seven patients. BMC Med Genet. 2016;17:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorlin JB, Yamin R, Egan S, et al. Human endothelial actin-binding protein (ABP-280, nonmuscle filamin): a molecular leaf spring. J Cell Biol. 1990;111:1089-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gurnett CA, Boehm S, Connolly A, Reimschisel T, Dobbs MB. Impact of congenital talipes equinovarus etiology on treatment outcomes. Dev Med Child Neurol. 2008;50:498-502. [DOI] [PubMed] [Google Scholar]
- 19.Jiang H, Liang S, He K, et al. Exome sequencing analysis identifies frequent oligogenic involvement and FLNB variants in adolescent idiopathic scoliosis. J Med Genet. 2020;57:405-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kircher M, Witten DM, Jain P, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genetics. 2014;46:310-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krakow D, Robertson SP, King LM, et al. Mutations in the gene encoding filamin B disrupt vertebral segmentation, joint formation and skeletogenesis. Nat Genet. 2004;36:405-410. [DOI] [PubMed] [Google Scholar]
- 22.Kruse LM, Dobbs MB, Gurnett CA. Polygenic threshold model with sex dimorphism in clubfoot inheritance: the Carter effect. J Bone Joint Surg Am. 2008;90:2688-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2019;25:1754-1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamura F, Stossel TP, Hartwig JH. The filamins: organizers of cell structure and function. Cell Adh Migr. 2011;5:160-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.National Center for Biotechnology Information. ClinVar; [VCV000599274.1]. Available at: https://www.ncbi.nlm.nih.gov/clinvar/variation/VCV000599274.1. Accessed March 29, 2020.
- 26.Online Mendelian Inheritance in Man (OMIM.) Available at: http://omim.org/. Accessed March 29, 2020.
- 27.Popowicz GM, Schleicher M, Noegel AA, Holak TA. Filamins: promiscuous organizers of the cytoskeleton. Trends Biochem Sci. 2006;31:411-419. [DOI] [PubMed] [Google Scholar]
- 28.Sadler B, Gurnett CA, Dobbs MB. The genetics of isolated and syndromic clubfoot. J Child Orthop. 2019;13:238-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shrimpton AE, Levinsohn EM, Yozawitz JM, et al. A HOX gene mutation in a family with isolated congenital vertical talus and Charcot-Marie-Tooth disease. Am J Hum Genet. 2004;75:92-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sung SS, Brassington AM, Grannatt K, et al. Mutations in genes encoding fast-twitch contractile proteins cause distal arthrogryposis syndromes. Am J Hum Genet. 2003;72:681-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Flier A, Kuikman I, Kramer D, et al. Different splice variants of filamin-B affect myogenesis, subcellular distribution, and determine binding to integrin [beta] subunits. J Cell Biol. 2002;156:361-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weymouth KS, Blanton SH, Bamshad MJ, et al. Variants in genes that encode muscle contractile proteins influence risk for isolated clubfoot. Am J Med Genet A. 2011;155:2170-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weymouth KS, Blanton SH, Powell T, Patel CV, Savill SA, Hecht JT. Functional assessment of clubfoot associated HOXA9, TPM1, and TPM2 variants suggests a potential gene regulation mechanism. Clin Orthop Relat Res. 2016;474:1726-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wynne-Davies R. Genetic and environmental factors in the etiology of talipes equinovarus. Clin Orthop Relat Res. 1972;84:9-13. [DOI] [PubMed] [Google Scholar]
- 35.Xu Q, Wu N, Cui L, et al. Comparative analysis of the two extremes of FLNB-mutated autosomal dominant disease spectrum: from clinical phenotypes to cellular and molecular findings. Am J Transl Res. 2018;10:1400-1412. [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Q Wu N Cui L Wu Z Qiu G. Filamin B: the next hotspot in skeletal research? J Genet Genomics. 2017;44:335-342. [DOI] [PubMed] [Google Scholar]
- 37.Yang CF, Wang CH, Siong H'ng W, et al. Filamin B loss-of-function mutation in dimerization domain causes autosomal-recessive spondylocarpotarsal synostosis syndrome with rib anomalies. Hum Mutat. 2017;38:540-547. [DOI] [PubMed] [Google Scholar]
- 38.Yang H, Zheng Z, Cai H, et al. Three novel missense mutations in the filamin B gene are associated with isolated congenital talipes equinovarus. Hum Genet. 2016;135:1181-1189. [DOI] [PubMed] [Google Scholar]
- 39.Zhou AX, Hartwig JH, Akyurek LM. Filamins in cell signaling, transcription and organ development. Trends Cell Biol. 2010;20:113-123. [DOI] [PubMed] [Google Scholar]