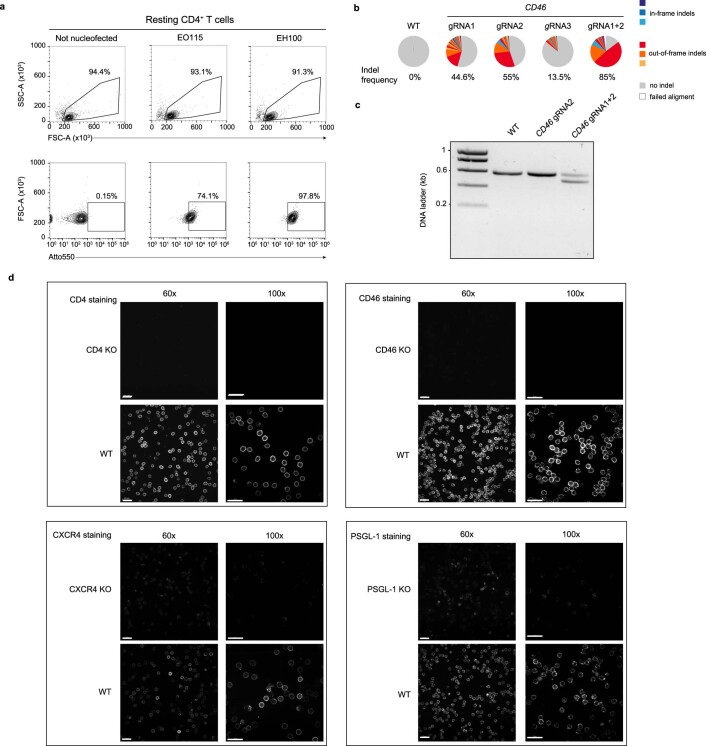
Extended Data Fig. 2. Optimized nucleofection allows RNP delivery into virtually every resting CD4+ T cell and results in efficient KO of single genes.
a, RNPs were generated by mixing crispr RNA (crRNA) together with a fluorescently labeled tracrRNA (Atto550; IDT) in an equimolar ratio and incubation at 95 °C for 5 min. The temperature was then decreased slowly to reach room temperature (around 1 hour). Next, the gRNA complex was mixed together with CAS9 for 15 min at room temperature, at molar ratios of 1:2.5 (Cas9:gRNA). Cells were mixed with the RNP complex and nucleofected in P3 buffer using either nucleofection program EH100 or EO115 (the latter suggested by the manufacturer). 24 h after nucleofection, the positivity for the fluorescent Atto5 label was analyzed by flow cytometry on a BD LSRFortessa. Cells mixed with the RNP fluorescent complex, but not nucleofected, served as control. FACS plots of one out of two experiments is shown. b,c Resting CD4+ T cells were nucleofected with RNPs specific for CD46 using either gRNA1, gRNA2, gRNA3 or a combination of gRNA1 + 2. One week later, cells were collected and lysed. A PCR specific for the CD46 locus was performed. The product was either analyzed by deep sequencing on an Illumina Miseq (b) or separated on an agarose gel (c). One experiment is shown (n = 2). d, Qualitative evaluation of the different gene KOs in resting CD4+ T cells by immunostaining and confocal microscopy two weeks after nucleofection. One representative donor each out of three is shown. Scale bars: 20 µm.