FIGURE 3.
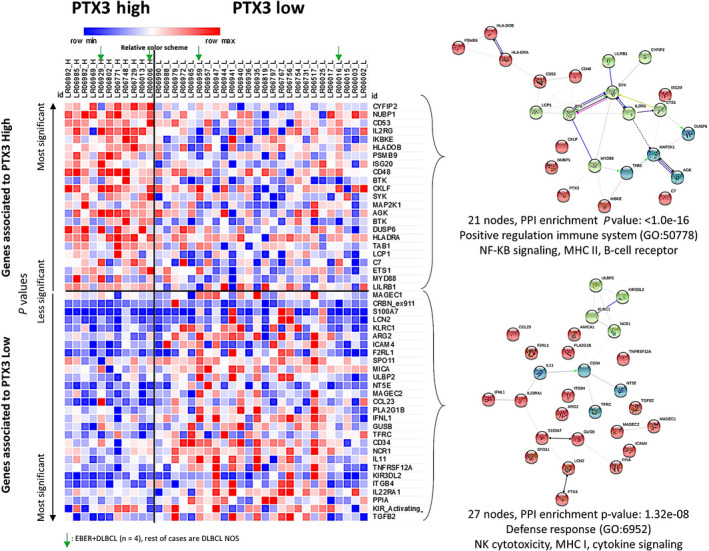
Gene expression analysis of the training set of patients with diffuse large B‐cell lymphoma (DLBCL). A set of 37 cases of DLBCL from Tokai University Hospital was selected and a gene expression analysis was carried out using Lymph2Cx and immuno‐oncology NanoString panels. Nonvariant genes between the two groups were excluded and significant genes were ranked according to P values and group associations. The heatmap visualization shows how gene expression between high and low pentraxin 3 (PTX3) groups differed, indicating a different pathogenic mechanism. A relative color scheme uses the minimum and maximum values in each row to convert values to colors and allows for the easier visualization of differences between the two groups. Genes involved in innate and adaptive immune responses and macrophage and Toll‐like receptor pathways, including CYFIP2, NUBP1, CD53, IL2RG, IKBKE, BTK, SYK, MAP2K1, and MYD88, were upregulated in the high PTX3 group. Genes involved in regulation, interleukins, adhesion, and natural killer (NK) cell function pathways, including TFFB2, KIR2DS1, PPIA, IL22RA1, and ITGB4, were upregulated in the low PTX3 group. Using the STRING database, the 21 genes associated with the high PTX3 group (top right panel) corresponded to the pathways positively regulating the immune system, nuclear factor‐κB signaling, MHC II, and B‐cell receptors. In contrast, the 27 genes associated with the low PTX3 group (bottom right panel) corresponded to the defense response pathway, NK cytotoxicity, MHC I, and cytokine signaling
