Abstract
Background
Prenatal alcohol exposure is the most common cause of birth defects and intellectual disabilities and can also increase the risk of stillbirth and negatively impact fetal growth.
Objective
The purpose of this study is to determine the effect of early prenatal alcohol exposure on non-human primate placental function and fetal growth. We hypothesize that early chronic prenatal alcohol will alter placental perfusion and oxygen availability that adversely effects fetal growth.
Study Design
Rhesus macaques self-administer 1.5g/kg/day ethanol (n=12) or isocaloric maltose-dextrin (n=12) daily preconception through the first 60 days of gestation (G60, term is G168). All animals were serially imaged with Doppler ultrasound to measure fetal biometry, uterine artery volume blood flow and placental volume blood flow. Following Doppler ultrasound, all animals also underwent both BOLD-MRI to characterize placental blood oxygenation and DCE-MRI to quantify maternal placental perfusion. Animals were delivered by cesarean section for placental collection and fetal necropsy at G85 (n=8), G110 (n=8), or G135 (n=8). Histologic and RNA-Seq analysis was performed on collected placental tissue.
Results
Placental volume blood flow was decreased at all gestational time points in ethanol-exposed vs. control, but most significantly at G110 by Doppler ultrasound (p<0.05). A significant decrease occurred in ethanol-exposed vs. control animals in total volumetric blood flow on DCE-MRI at both G110 and G135 (p<0.05) as well as a global reduction in T2*, high blood deoxyhemoglobin concentration, throughout gestation (p<0.05). Similarly, evidence of placental ischemic injury was notable by histologic analysis, which revealed a significant increase in microscopic infarctions in ethanol-exposed, not control, largely present at mid to late gestation. Fetal biometry and weight were decreased in ethanol-exposed vs. control, but not significant. Analysis with RNA-Seq suggests involvement of the inflammatory and extracellular matrix response pathways.
Conclusions
Early chronic prenatal alcohol exposure significantly diminishes placental perfusion at mid- and late-gestation and significantly decreases the oxygen supply to the fetal vasculature throughout pregnancy, associated with microscopic placental infarctions in the non-human primate. Although placental adaptations may compensate for early environmental perturbations to fetal growth, placental blood flow and oxygenation is reduced, consistent with evidence of placental ischemic injury.
Keywords: Fetal alcohol spectrum disorder, fetal alcohol syndrome, maternal drinking, placental perfusion and oxygenation, prenatal alcohol exposure
CONDENSATION:
First trimester alcohol exposure alters placental function and fetal growth in a rhesus macaque model
INTRODUCTION
Alcohol freely crosses the placenta and can accumulate in the fetus at levels comparable to maternal blood alcohol concentrations.1 Prenatal alcohol exposure increases the risk of preterm birth, stillbirth, decreased fetal growth and fetal alcohol spectrum disorder (FASD), the most common non-genetic cause of cognitive impairment in the United States.2,3 Currently, there are no approved drugs to treat FASD or established tools to prevent adverse outcomes. Among pregnant women in the United States, approximately 10% have consumed any alcohol the past 30 days,4 resulting in greater than three quarters of a million alcohol-exposed fetuses.5-7
The placenta occupies a central role in supporting normal fetal growth and development during pregnancy.8 Prior studies have suggested that placental dysfunction may contribute to intrauterine growth restriction (IUGR) in FASD,2 but the mechanisms and specific vasoactive effects of alcohol linking placental dysfunction to disrupted fetal growth remains an area of ongoing scientific exploration. Antenatal ethanol exposure has been previously shown to induce apoptosis in human placental trophoblast cells, disrupt trophoblast cell motility, and potentially affect uterine spiral artery remodeling.9-13 Prior pregnant animal studies using ovine14 and baboon15-18 models have demonstrated abnormal uterine and cerebral blood flow following acute ethanol exposure and in a rat model impaired uterine artery vasodilation from chronic binge drinking.19
Non-human primate (NHP) fetal ontogeny, placental structure, and ethanol absorption and metabolism more closely resembles that of humans than other animal models.17 We recently developed a NHP model of first trimester ethanol consumption that generated pregnancies from control animals and dams that drank 1.5g/kg of ethanol daily (~6 standard drinks for humans).21 Fetal brain maturation was characterized with in utero magnetic resonance imaging (MRI) and ex vivo slice electrophysiology, which revealed that cerebellar and brainstem fetal growth is diminished, in this model of FASD22 Moreover, fetal diffusion MRI indicated altered maturation of motor-related white matter fiber systems, and these findings were corroborated with altered synaptic development in cortical and striatal regions found by electrophysiology22. Previously, a pilot investigation that demonstrated feasibility and greater sensitivity of in utero MRI directed at the placenta to detect the effects of early-pregnancy drinking on placental function and fetal development23 over standard clinical doppler-ultrasound (Doppler-US). Placental blood flow was measured using dynamic contrast-enhanced MRI (DCE-MRI)24 and oxygen exchange was quantified through analysis of water T2* values via the blood oxygen level-dependent (BOLD) effect.25
Utilizing these in vivo MRI methods at gestational day 110 (G110, term is ~168 days) and G135, we observed that early gestation alcohol exposure decreased both placental perfusion and fetal oxygen supply mid-gestation and was associated with both decreased fetal and brain weight.23 However, the underlying mechanisms contributing to the observed altered placental function and fetal development were not explored in this prior work. The primary objective of this study was to utilize the complete set of animals in which placental tissue is available, including at an additional gestational timepoint (G85), to evaluate the adverse effects of early, chronic prenatal alcohol exposure on placental outcomes and fetal growth. Our second objective was to study the effects of prenatal ethanol exposure on placental histology and gene expression to identify mechanisms underlying placental dysfunction detectable using non-invasive imaging approaches.
MATERIALS AND METHODS
Experimental Design
All protocols were approved by the Institutional Animal Care and Use Committee of the Oregon National Primate Research Center, and guidelines for humane animal care were followed. The generation of pregnancies in the NHP FASD model has been previously published.21 This study focuses on the subset of time-mated pregnant rhesus macaques (n=24) consisting of 12 control and 12 ethanol-exposed animals that underwent placental collection at time of delivery. Dams were trained to orally self-administer daily either 1.5g/kg/day of 4% ethanol solution (~6 drinks/day) or an isocaloric control fluid with training being initiated at least 4 months prior to undergoing time-mated breeding with plasma estradiol sampling26-28 (Figure 1). The day of peak plasma estradiol was defined as gestational day 0 (G0). Each pregnant animal continued daily drinking of 1.5 g/kg/day ethanol ad lib until G60, at which point access to ethanol, or isocaloric solution, was removed (Figure 1). All animals underwent Doppler-US followed by MRI consisting of T2 and DCE measurements and after imaging, immediate cesarean section delivery with placenta collection and fetal necropsy at G85 (n=8), G110 (n=8) or G135 (n=8) (Figure 1). Collected placental tissue was processed in liquid nitrogen for molecular analysis, RNAlater for RNA-Seq, and formalin fixation for histology.
Figure 1. Study Design Overview.
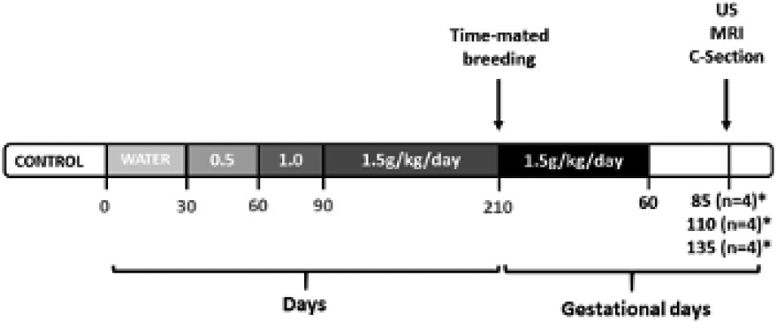
Timeline of the experimental design indicating that the ethanol-exposed animals were trained to self-administer ethanol with incremental dosing increase until reaching a dose of 1.5g/kg/day (~6 drinks/day) which was then maintained through the first 60 days post-conception before being discontinued. The age-matched control group (*) self-administered an isocaloric control fluid also through gestational day 60 (G60, term is ~168 days). Ultrasound and MRI were performed at G85, G110 and G135 with immediate cesarean delivery following imaging at G135.
Imaging
Doppler-US
All ultrasounds were performed by a single sonographer (J.O.L) using image-directed pulsed and color Doppler equipment (GE Voluson 730) with a 5- to 9-MHz sector probe. Animals were sedated by intramuscular administration of 10 mg/kg ketamine (Henry Schein Animal Health®) and maintained on a portable anesthesia delivery system providing O2 with 1.5% isoflurane. Doppler waveform measurements for the uterine artery (Uta) and umbilical artery were performed using machine-specific software. The following measurements were obtained: pulsatility index (PI), velocity time integral (VTI), and fetal heart rate (HR) to calculate uterine artery blood flow (cQUta) and placental volume blood flow (cQUV) as previously described.23,29-33 cQUta was calculated and corrected by maternal weight as: cQUta= VTI x CSA (Uta cross-sectional area) x HR. 29-33 Placental volume blood flow (cQUV) was calculated as: mean velocity (Vmean) x CSA x 60. 29-33
Placental MRI
Immediately following ultrasound, MRI studies were performed on a 3T Siemens TIM-Trio scanner (Erlangen, Germany) as previously published.23 Following localization of the placenta and acquisition of anatomic images in the coronal and axial planes, axial 2D multislice spoiled multiecho gradient echo images (TR=418 ms, flip angle=30°, 256x72 matrix, 96 slices, 1.5 mm isotropic spatial resolution) spanning the entire uterus were acquired at six in-phase echo times (TE=4.92, 9.84, 19.68, 29.52, 36.90, and 44.28 ms) for T2* measurements, and T1 was measured with the variable flip angle (VFA) method.34 After acquisition of VFA data, 150 3D SPGR images were acquired for DCE-MRI (TR=2.00 ms, TE=0.72 ms, flip angle=20°, acquisition time of 3.64 seconds), with field of view and resolution matched to the VFA images. Ten baseline images were acquired prior to intravenous injection of 0.1 mmol/kg of gadoteridol contrast reagent (Prohance®, Bracco Diagnostics Inc, Princeton, NJ) at a rate of 30 mL/min using a syringe pump (Harvard Apparatus, Holliston, MA). Anatomic and multiecho imaging was performed during expiratory breath holding, while DCE-MRI data were acquired during ventilated breathing. Physiological monitoring of pulse rate, arterial blood oxygen saturation, and end-tidal CO2 partial pressure was performed throughout, with no deviations from normal ranges observed in these parameters. BOLD and DCE-MRI analyses were performed as previously described.23,34,35
Placental histology
Formalin fixed paraffin-embedded histologic sections were stained with hematoxylin and eosin and reviewed by a single placental pathologist (T.K.M.) blinded to exposure and outcomes. Tissue sections were scored for any signs of infection and classic histologic features of maternal vascular mal-perfusion, including infarctions and/or accelerated villous maturation.36
Statistical Analysis
Data are expressed as mean ± SD. All animals (n=24) were analyzed and differences between ethanol-exposed and controls at G85, G110 and G135 were tested by a 2-way ANOVA with post-hoc Tukey comparison, significance was designated at p<0.05. The T2* results were evaluated using a two-sample Kolmogorov-Smirnov test.
Gene Expression
RNA Isolation and Quality Assessment
Dissected placenta tissue samples (n=24) in RNAlater (ThermoFisher Scientific) were delivered to the OHSU Gene Profiling Shared Resource, where phenol-chloroform extraction was performed followed by RNA isolation using the RNeasy Mini kit (QIAGEN). RNA integrity and size distribution were assessed using a 2100 Bioanalyzer (Agilent Technologies). Four samples were excluded due to suboptimal RNA yield.
RNA Sequencing
RNA-seq libraries were prepared using the TruSeq with Ribosomal Depletion kit (Illumina). The amplified product was profiled on the TapeStation (Agilent). Libraries were quantified using real time PCR (Kapa Biosystems) and run on a HiSeq 2500 (Illumina). The resulting base call files were converted to fastq files using bcl2fastq (Illumina)
Gene-level Differential Expression Analysis
Differential expression analysis utilized standard operating procedures established by the ONPRC Bioinformatics & Biostatistics Core. The quality of the raw sequencing files was evaluated using FastQC37 combined with MultiQC (http://multiqc.info/).38 Trimmomatic39 was used to remove any remaining Illumina adapters. Reads were aligned to Ensembl’s mmul8 along with its corresponding annotation, release 87. The program STAR40 (v2.7.3a) was used to align the reads to the genome and RNA-SeQC41 was utilized to ensure alignments were of sufficient quality.
The differential expression analysis was performed in the open-source software R.42 Gene-level raw counts were filtered to remove genes with extremely low counts in many samples following the published guidelines,43 normalized using the trimmed mean of M-values method (TMM),44 and transformed to log-counts per million with associated observational precision weights using voom45 method. Gene-wise linear models with primary variables treatment group, gestational stage and their interaction, adjusting for sex and technical factors (RNA processing and sequencing batch)were employed for differential expression analyses using limma with empirical Bayes moderation46 and false discovery rate (FDR) adjustment.47
RNA Pathway Analysis
A total of 12,879 genes were considered expressed in the 18 placental samples dataset after filtering. To adjust for multiple comparisons, an FDR adjusted p-value of < 0.2 was used to test for significance because of the small sample size and degree of biological variation. As is common in longitudinal NHP studies with relatively high variability and small group sizes, few genes were identified as differentially expressed at FDR < 0.2 in the comparisons between ethanol and control or comparisons between gestational time points, except in ethanol-exposed samples at G135 vs G85. To overcome this limitation, pathway-based patterns were explored to look for effects on gene networks, pathways, and biological processes due to maternal ethanol consumption. Pathway analysis48 was performed in Ingenuity Pathway Analysis (IPA). (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis). The core analysis option in IPA was used to perform pathway analysis using an unadjusted p-value < 0.05 and fold change > 1.5 for all of the pairwise post-hoc comparisons.
RESULTS
Growth parameters
Ultrasound measurements of fetal biometry were not significantly different between fetuses exposed to ethanol and controls at all gestational ages (Table 1). There was no significant difference in fetal birth weight at time of delivery in ethanol-exposed fetuses versus control animals at G85 (p=0.5), G110 (p=0.07) and G135 (p=0.1) (Table 2). Maternal weights and fetal gender ratios were not significantly different across treatment groups (Table 2). All major fetal organs were examined by a veterinary pathologist and no gross structural anomalies were noted in ethanol-exposed pregnancies.
Table 1.
Fetal biometry
| Parameter | Gestational day 85 | Gestational day 110 | Gestational day 135 | |||
|---|---|---|---|---|---|---|
| Control (n=4) |
Ethanol (n=4) |
Control (n=4) |
Ethanol (n=4) |
Control (n=4) |
Ethanol (n=4) |
|
| BPD (mm) | 29.4 ± 1.4 | 28.3 ± 1.6 | 38 ± 1.7 | 36.7 ± 3.5 | 44.5 ± 0.8 | 42.6 ± 2.6 |
| AC (mm) | 9 ± 0.3 | 8.9 ± 0.5 | 12 ± 0.4 | 11.6 ± 0.7 | 13.7 ± 0.4 | 12.9 ± 1.4 |
| FL (mm) | 17.9 ± 1.2 | 6.8 ± 0.9 | 28.5 ± 0.9 | 27.3 ± 1.2 | 36.6 ± 1.9 | 36 ± 0.4 |
Definition of abbreviations:
BPD = biparietal diameter
AC = abdominal circumference
FL = femur length
2-Way ANOVA used with post-hoc Tukey comparison test. Data are means ± SD.
p<0.05
Table 2.
Maternal, fetal birth, and placental weights
| Parameter | Gestational day 85 | Gestational day 110 | Gestational day 135 | |||
|---|---|---|---|---|---|---|
| Control (n=4) |
Ethanol (n=4) |
Control (n=4) |
Ethanol (n=4) |
Control (n=4) |
Ethanol (n=4) |
|
| Maternal weight (kg) | 7.1±0.8 | 6.8±0.9 | 8.1±1.9 | 7.8±1.5 | 7.8±1 | 8.3±0.9 |
| Fetal weight (g) | 81.7±7.8 | 77.8±10.3 | 209±23.8 | 183±16.7 | 334±9.5 | 323±17 |
| Placental weight (g) | 49.4±15 | 42.9±3.6 | 77±14.7 | 64.6±4.1 | 81±6.2 | 82.4±8.6 |
| Fetal sex (male:female) | 3:1 | 2:2 | 1:3 | 1:3 | 1:3 | 1:3 |
2-Way ANOVA used with post-hoc Tukey comparison test. Data are means ± SD.
p<0.05
Placental perfusion and oxygenation
Both cQuta and cQuv was smaller in the ethanol-exposed group compared to controls at G110 (p<0.05), but differences were not seen at G85 or G135 (Table 3). Uterine artery pulsatility indices were increased in ethanol-exposed animals, but was not statistically significant at all gestational time points (Table 3). Similarly, there was a trend of increased umbilical artery pulsatility indices suggestive of increased placental vascular impedance across all time points that was significant at G85 (p<0.05) (Table 3).
Table 3.
Doppler ultrasound and Dynamic contrast-enhanced MRI measurements of placental function and oxygenation
| Parameter | Gestational day 85 | Gestational day 110 | Gestational day 135 | |||
|---|---|---|---|---|---|---|
| Control (n=4) |
Ethanol (n=4) |
Control (n=4) |
Ethanol (n=4) |
Control (n=4) |
Ethanol (n=4) |
|
| Uterine artery PI | 0.7±0.1 | 0.9±0.2 | 0.6±0.1 | 0.8±0.1 | 0.8±0.3 | 0.85±0.1 |
| Umbilical artery PI | 1.7±0.3 | 2.4±0.3* | 1.3±0.2 | 1.6±0.3 | 1.1±0.06 | 1.4±0.3 |
| cQuta (ml/min/kg) | 22.3±7.3 | 14±6.3 | 41±23 | 22±11.5* | 26.5±14 | 26±1.4 |
| cQuv | 17.7±2.2 | 9.8±3.6 | 24.6±4.8 | 15.1±0.7* | 31.5±7.1 | 28.9±13.7 |
| Total Placental blood flow (ml/min)§ | 289±120 | 121±27 | 681±39.6 | 285±142* | 559±190 | 414±221* |
Definition of abbreviations:
= obtained by DCE-MRI
PI = pulsatility index, VTI = velocity time integral
CSA (cross section of uterine artery) = π(diameter/2)2
Vmean (mean velocity) = 0.5 x maximum umbilical vein velocity
cQuta (uterine artery blood flow) = VTI x CSA x HR adjusted for maternal weight
cQuv (placental volume blood flow) = Vmean x CSA x 60
2-Way ANOVA used with post-hoc Tukey comparison test. Data are means ± SD.
p<0.05
Maternal perfusion of the placental intervillous space was evaluated using MRI and multiple irregularities were seen in ethanol-exposed animals compared to controls. Using DCE-MRI, comprehensive maternal perfusion through spiral arteries is quantified as total volumetric blood flow. This was found to be significantly lower (p<0.05) at G110 and G135 in ethanol-exposed group versus controls (Table 3), consistent with the semi-quantitative measures of cQuta at G110.
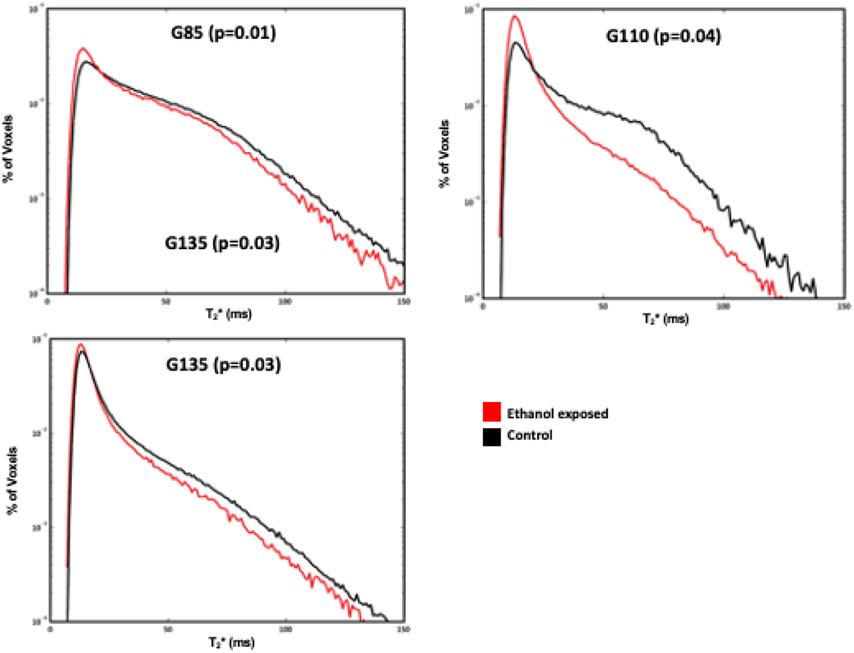
Placental oxygen supply was assessed through analysis of water T2 values. In control placentas at all three timepoints, MR image voxels proximal to spiral artery sources of oxygenated maternal blood are characterized by relatively long T2 (Figure 2), as previously described.35 At greater distances from the spiral arteries, the concentration of deoxyhemoglobin is higher, secondary to fetal oxygen uptake, resulting in decreased T2*. The histograms shown in Figure 2 summarize placental T2*, demonstrating a statistically significant reduction in T2* across gestation, but most prominently at G85 (p=0.01) compared with G110 (p=0.04) and G135 (p=0.03) in the ethanol-exposed cases compared to controls.
Figure 2. Histogram plot of T2* versus percent of placental voxels displayed for ethanol-exposed (red) vs. control animals (black) at G85, G110 and G135.
Ethanol-exposed animals had a smaller fraction of large T2* values compared to controls across all time points, demonstrating decreased fetal oxygen availability in the former.
Placental histology
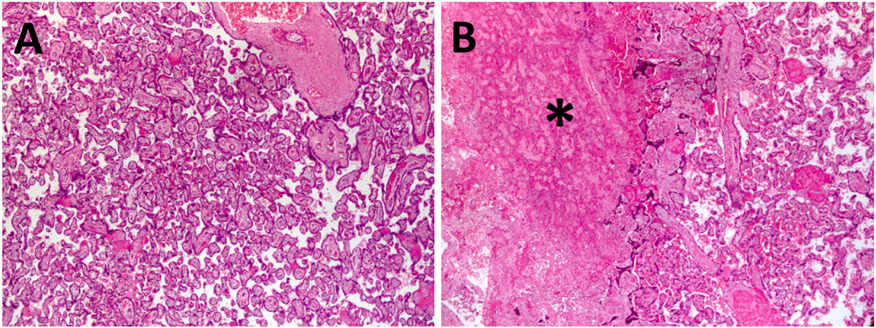
Placental pathology demonstrated increased frequency of microscopic (<1.0cm) infarctions in placentas exposed to ethanol (5/12, p<0.05) compared with controls (0/12) (Figure 3). These infarctions were predominantly seen at G110 (2/4) and G135 (2/4) (Figure 3). Larger placental infarctions were also seen in a few cases, but only in ethanol-exposed placentas (Figure 3). There was no histologic evidence of infection, increase in placental villi maturation or findings of chorangiosis. Placental weights were not different amongst different treatment groups at all three pregnancy timepoints (Table 2).
Figure 3. Placental histologic changes associated with early maternal ethanol exposure.
(A) Representative control H&E stained placental section at G110 compared with microscopic infarctions (asterix) seen in (B) ethanol-exposed animals. Magnification is 50x.
RNA Sequencing
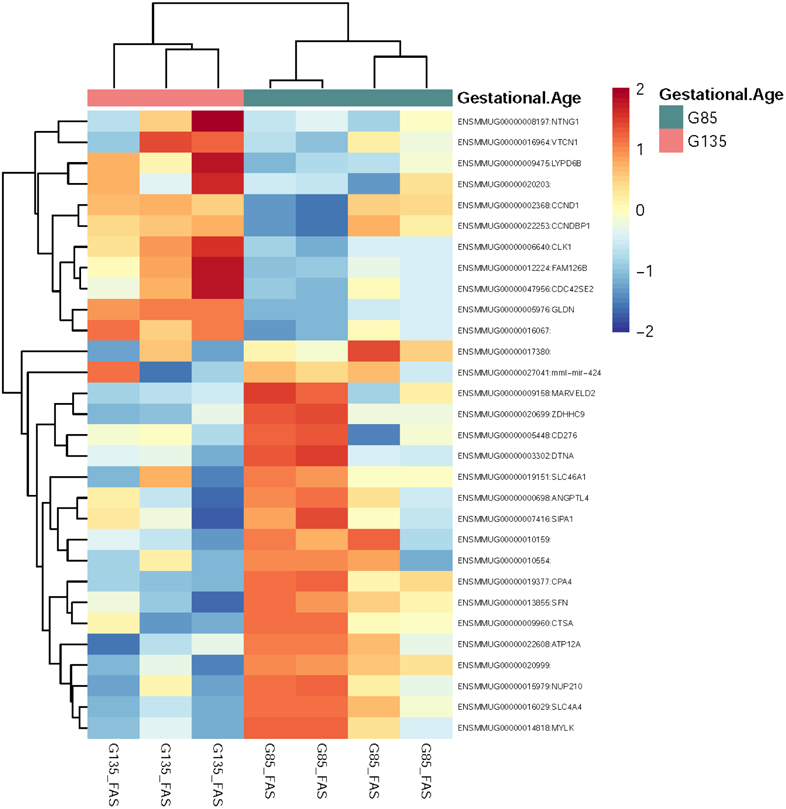
Pairwise comparisons between sample groups were performed, and the comparison with the most significantly differentially expressed genes was G135 vs. G85 in the ethanol-exposed samples: 505 upregulated genes were identified in G135 compared to 397 upregulated genes in G85 (FDR<0.2) (Figure 4). For the pathway analysis of G135 vs. G85 in ethanol-exposed placentas, there were 1356 analysis-ready molecules with p-value < 0.05 and fold-change > 1.5. IPA identified 71 significant pathways. Of note, Ethanol Degradation IV, Oxidative Ethanol Degradation III, and Ethanol Degradation II pathways have negative z-scores indicating predicted inhibition of these pathways at G135 vs. G85 in ethanol-exposed samples but not controls.
Figure 4. RNA Seq Heatmap.
Heatmap of top 50 differentially expressed genes by FDR adjusted p-value between FAS G135 and G85.
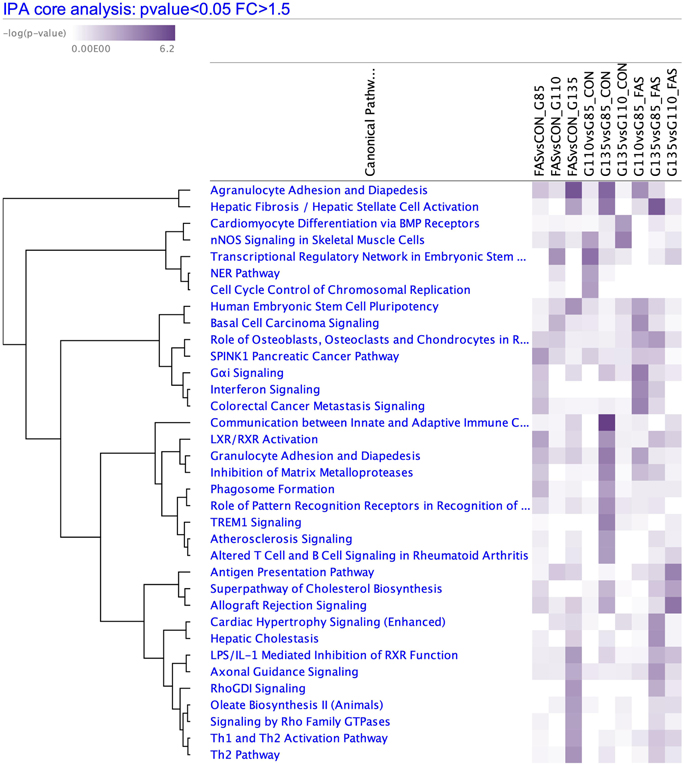
Comparing ethanol-exposed samples to control samples at each timepoint, pathway analysis was used to look for patterns affecting multiple genes of related function, utilizing thresholds of unadjusted p-value < 0.05 and fold change > 1.5. Pathways with p < 0.001 are shown in Figure 5. Many of these contain genes involved with extracellular matrix remodeling and inflammation.
Figure 5. RNA Seq Pathway Analysis.
Significant pathways (p<0.001) when comparing ethanol-exposed (FAS) and control (CON) samples at each timepoint and between timepoints within each treatment group.
COMMENT
Principal Findings
Ethanol exposure through the first 60 days post-conception, akin to the first trimester in human pregnancies, reduced both placental blood flow and oxygenation, with more dramatic effects on placental perfusion at G110 and G135 (Table 3). These findings were accompanied by increased placental pathology and associated with differences in the expression of genes related to inflammation and maintenance of the extracellular matrix. Additionally, ethanol-exposed placentas demonstrated inhibition of gene pathways related to ethanol degradation at G135 compared to G85, not seen in control placentas.
Clinical Implications
Ethanol readily crosses the placenta and accumulates in the fetus at concentrations proportionate to maternal blood levels within an hour.1 Consistent with our earlier study21 and existing literature,49 we have shown in a larger cohort that early, chronic prenatal alcohol exposure plays an important role in altered placental perfusion and function. These findings suggest that placental impairment secondary to early prenatal ethanol exposure is a possible contributor to the previously reported altered fetal brain development22 and decreased cerebral artery blood flow16-18 since the placenta provides oxygen and transit of essential metabolites for routine fetal nervous system development.50
Our analysis by RNA-Seq suggests involvement of inflammatory and extracellular matrix response pathways contributing to the observed decreased placental perfusion and fetal oxygen availability. These findings played a role in the significant presence of placental microinfarctions, suggestive of reduced functional villi, in ethanol-exposed animals only. Early ethanol exposure may have altered these pathways and contributed to endothelial dysfunction in the placental vasculature. Similarly, in a NHP model of Western Style diet (WSD), we had found that consumption of a WSD during pregnancy exacerbated placental inflammation, reduced uterine blood flow and significantly increased the frequency of both placental infarctions and stillbirth.29 Additionally, RNA-Seq findings of downregulated Ethanol Degradation II, III, IV genes at G135 compared to G85 in ethanol-exposed placentas, but not controls suggests ethanol metabolism pathway inhibition. This is possibly another adaptive placental response to early-ethanol exposure to protect against ethanol toxicity. Other studies have also similarly reported altered ethanol degradation pathways in gestational alcohol exposure.51
Decreased placental perfusion and fetal oxygen availability occurred most significantly at mid- to late-gestation, but was lowest at G110, 50 days after ethanol cessation. Although in a smaller subset of animals, early alcohol exposure appeared to impact fetal birth weight23, in this larger cohort, preserved fetal growth and placental weight was observed, suggestive of the NHP placenta’s adaptive capabilities to early transient environmental perturbations19,52 when compared with chronic prenatal insults throughout gestation.25,53,54 In spite of this compensatory response, the reduced placental oxygenation at G135 combined with the presence of placental microinfarctions suggest persistent placental injury that could further disrupt fetal development in ongoing pregnancies.
Research Implications
The placenta has significant functional reserve to maintain adequate fetal nutrient delivery during pregnancy. However, when a critical threshold for placental compensation is reached,23,53 it can result in complications such as fetal growth restriction with increased fetal and neonatal mortality and morbidity.19 Thus, it is important to determine a non-invasive method to identify pregnancies at risk of hemodynamic alterations before the fetus is affected.
RNA-Seq was also performed to examine gene expression differences suggestive of pathways important to placental development and function that are affected by early chronic ethanol exposure. Our findings suggestive of inflammatory and extracellular matrix remodeling pathway involvement reveal mechanistic targets for future investigation for early intervention in pregnancies affected by first trimester alcohol exposure prior to pregnancy detection.
Strengths and Limitations
This study builds upon our prior earlier work23 by using a larger cohort to investigate of the biological effects of early prenatal alcohol exposure on placental function and fetal growth. Also, by adding a gestational time point and utilizing placental histology and gene expression analysis, we were able to establish correspondences between in vivo imaging findings and identified placental pathology and RNA-Seq results.
This animal model provides precise control over experimental variables such as gestational age and quantity and timing of ethanol exposure throughout gestation. Uncertainties in such variables confound studies of human subjects. All placentas were collected at time of cesarean section delivery prior to onset of spontaneous labor from pregnancies with similar environmental exposures, including diet. Thus, the direct effects of early alcohol consumption were more readily assessed compared to human studies with weaker correlative measures.
Limitations of this study were the animal cohort size and cross-sectional study design, chosen to avoid confounds associated with repeated isoflurane exposure, used for sedation during MRI procedures. Future studies from our group will focus on longitudinal assessments within the same animal to allow for more direct interrogation of this placental acclimatization phenomenon. Additionally, with RNA Seq analysis our sample size did not provide the power to detect differentially expressed genes with small effect sizes between the experimental groups at a single timepoint.
Conclusions
Early chronic prenatal ethanol exposure significantly impairs maternal placental perfusion and oxygen supply to the fetal vasculature throughout pregnancy resulting in associated placental pathology in voluntary-drinking rhesus macaques. These alterations are detectable early in gestation using newly developed non-invasive imaging techniques, demonstrating increased sensitivity over standard Doppler-US. Abnormal gene expression patterns related to inflammation and the extracellular matrix are associated with the observed functional deficits. Placental adaptations were able to compensate for the adverse effect to placental blood flow and oxygenation by maintaining normal fetal growth through G135 in this experimental cohort. At this time, there is no treatment for FASD or the adverse fetal affects from prenatal alcohol exposure. Thus, it is important for women thinking of conceiving or those who are pregnant to adhere to the recommendations by ACOG to abstain from alcohol consumption.
AJOG AT A GLANCE.
A. Why was the study conducted?
This study was conducted to determine the effects of chronic maternal first trimester binge drinking on placental function and fetal growth.
B. What are the key findings?
Early chronic prenatal alcohol exposure significantly diminishes placental perfusion at mid- and late-gestation and significantly decreases the oxygen supply to the fetal vasculature throughout pregnancy in the rhesus macaque.
C. What does this study add to what is already known?
This study established correspondences between reduced comprehensive placental blood flow and oxygen availability on in-vivo MRI, increased placental microinfarctions on histology and upregulated inflammatory and extracellular matrix remodeling pathways by RNA-Seq in pregnancies with early alcohol exposure.
ACKNOWLEDGEMENTS
Financial Support: All Oregon National Primate Research Center (ONPRC) cores and units were supported by the National Institutes of Health (NIH) Grant P51 OD011092. Research reported in this publication was supported by the Reproductive Scientist Development Program, March of Dimes, and Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) under Award Numbers K12 HD000849 (to J.L.), R01 AA021981 (to C.K.) and R01 HD086331 (to A.F). The contents of this study are solely the responsibility of the authors and do not necessarily represent the official view of NIH NICHD and NIAAA.
We would also like to thank the veterinarian and husbandry staff at the ONPRC who provided excellent care for the animals used in this study.
Footnotes
Conflict of Interest Statement: None of the authors have financial or other relationships that could result in a conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Presented orally at the International Society of Magnetic Resonance Medicine Workshop on MRI of the Placenta in Atlanta, Georgia, February 4-5th, 2018 and as a poster at the 65th Society of Reproductive Investigation Annual Meeting in San Diego, California, March 6-10th, 2018.
REFERENCES:
- 1.Idanpaan-Heikkila J, Jouppila P, Akerblom HK, Isoaho R, Kauppila E, Koivisto M. Elimination and metabolic effects of ethanol in mother, fetus, and newborn infant. Am J Obstet Gynecol. 1972;112:387–393. [DOI] [PubMed] [Google Scholar]
- 2.O’Leary CM, Nassar N, Kurinczuk JJ, Bower C. The effect of maternal alcohol consumption on fetal growth. BJOG. 2009;116(3):390–400. [DOI] [PubMed] [Google Scholar]
- 3.Hannigan JH, Armant DR. Alcohol in pregnancy and neonatal outcome. Semin Neo. 2000;5(3):243–54. [DOI] [PubMed] [Google Scholar]
- 4.Tan CH, Denny CH, Cheal NE, Sniezek JE, Kanny D. Alcohol use and binge drinking among women of childbearing age – United States, 2011-2013. MMWR Morb Mortal Wkly Rep. 2015;64(37):1042–1046. [DOI] [PubMed] [Google Scholar]
- 5.Alcohol and Pregnancy. Centers for Disease Control and Prevention; website: http://www.cdc.gov/vitalsigns/fasd/. Updated February 2, 2016. Last accessed January 26, 2021. [Google Scholar]
- 6.Edwards EM, Werler MM. Alcohol consumption and time to recognition of pregnancy. Matern Child Health J. 2006;10(6):467–472. [DOI] [PubMed] [Google Scholar]
- 7.Floyd RL, Decoufle P, Hungerford DW. Alcohol use prior to pregnancy recognition. Am J Prev Med. 1999; 17(2):101–107. [DOI] [PubMed] [Google Scholar]
- 8.ACOG Practice Bulletin No. 134: Fetal growth restriction. Obstet Gynecol 2013; 121(5):1122–1133. [DOI] [PubMed] [Google Scholar]
- 9.Gundogan F, Elwood G, Longato L, et al. Impaired placentation in fetal alcohol syndrome. Placenta. 2008;29(2):148–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gundogan F, Elwood G, Mark P, et al. Ethanol-induced oxidative stress and mitochondrial dysfunction in rat placenta: relevance to pregnancy loss. Alcoholism, Clinical and Experimental Research. 2010;34(3):415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gundogan F, Gillican J, Qi W, Chen E, NAram R, de la Monte SM. Dose effect of gestational ethanol exposure on placentation and fetal growth. Placenta. 2015;36(5):523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siler-Khodr TM, Yang Y, Grayson MH, Henderson GI, Lee M, Schenker S. Effect of ethanol on thromboxane and prostacyclin production in the human placenta. Alcohol. 2000;21(2):169–180. [DOI] [PubMed] [Google Scholar]
- 13.Taylor SM, Heron AE, Cannell GR, Florin TH. Pressor effect of ethanol in the isolated perfused human placental lobule. Eur J Pharmacol. 1994;270(4):371–374. [DOI] [PubMed] [Google Scholar]
- 14.Falconer J The effect of maternal ethanol infusion on placental blood flow and fetal glucose metabolism in sheep. Alcohol Alcohol. 1990;25(4):413–416. [PubMed] [Google Scholar]
- 15.Kochunov P, Castro C, Davis D, et al. Fetal brain during a binge drinking episode: A dynamic susceptibility contrast MRI fetal brain perfusion study. Neuroreport. 2010;21(10):716–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.North K, Tobiasz A, Sullivan RD, et al. Prenatal alcohol exposure, anesthesia, and fetal loss in baboon model of pregnancy. J Drug Alcohol Res. 2019;7:236064. [PMC free article] [PubMed] [Google Scholar]
- 17.Simakova M, Tobiasz A, Sullivan RD, et al. Gestational age-dependent interplay between endocannabinoid receptors and alcohol in fetal cerebral arteries. J Drug Alcohol Res. 2019;8:236068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tobiasz AM, Duncan JR, Bursac Z, et al. The effect of prenatal alcohol exposure on fetal growth and cardiovascular parameters in a baboon model of pregnancy. Repro Sci. 2018;25(7):1116–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian K, Naik VD, Sathishkumar K, et al. Chronic binge alcohol exposure during pregnancy impairs rat maternal uterine vascular function. Alcohol Clin Exp Res. 2014;38(7):1832–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green KL, Szeliga KT, Bowen CA, Kautz MA, Azarov AV, Grant KA. Comparison of ethanol metabolism in male and female cynomolgus macaques (Macaca fascicularis). Alcohol Clin Exp Res. 1999;23(4):611–616. [PubMed] [Google Scholar]
- 21.Jimenez VA, Wang X, Newman N, et al. Detecting neurodevelopmental effects of early-gestation ethanol exposure: a nonhuman primate model of ethanol drinking during pregnancy. Alcohol Clin Exp Res. 2019;43(2):250–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Cuzon Carlson VC, Studholme C, et al. In utero MRI identifies consequences of early-gestation alcohol drinking on fetal brain development in rhesus macaques. Proc Natl Acad Sci USA. 2020;117(18):10035–10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lo JO, Schabel MC, Roberts VHJ, et al. First trimester alcohol exposure alters placental perfusion and fetal oxygen availability affecting fetal growth and development in a non-human primate model. Am J Obstet Gynecol. 2017;216(3):302.e1–302.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frias AE, Schabel MC, Roberts V, et al. Using dynamic contrast enhanced MRI to quantitatively characterize maternal vascular organization in the primate placenta. Magn Reson Med. 2015;73(4):1570–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schabel MC, Roberts VHJ, Lo JO, et al. Functional imaging of the non-human primate placenta with endogenous blood oxygen level-dependent contrast. Magn Reson Med. 2016;76(5):1551–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grant KA, Leng X, Green HL, Szeliga KT, Rogers LS, Gonzales SW. Drinking typography established by scheduled induction predicts chronic heavy drinking in a monkey model of ethanol self-administration. Alcohol Clin. Exp. Res, 2008;32(10):1824–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vivian JA, Green HL, Young JE, et al. Induction and maintenance of ethanol self-administration in cynomolgus monkeys (Macaca fascicularis): long-term characterization of sex and individual differences. Alcohol Clin Exp Res. 2001;25(8):1087–1097. [PubMed] [Google Scholar]
- 28.Falk JL. Production of polydipsia in normal rats by an intermittent food schedule. Science. 1961;133(3447):195–196. [DOI] [PubMed] [Google Scholar]
- 29.Frias A, Morgan T, Evans A, et al. Maternal high-fat diet disturbs uteroplacental hemodynamics and increases the frequency of stillbirth in a nonhuman primate model of excess nutrition. Endo. 2011;152(6):2456–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konje JC, Kaufmann P, Bell SC, Taylor DJ. A longitudinal study of quantitative uterine blood flow with the use of color power angiography in appropriate for gestational age pregnancies. Am J Obstet Gynecol. 2001;185(3):608–613. [DOI] [PubMed] [Google Scholar]
- 31.Acharya G, Sitras V, Erkinaro T, et al. Experimental validation of uterine artery volume blood flow measurements by Doppler ultrasonography in pregnant sheep. Ultrasound Obstet Gynecol. 2007;29(4):401–406. [DOI] [PubMed] [Google Scholar]
- 32.Acharya G, Wilsgaard T, Berntsen GK, Maltau JM, Kiserud T. Doppler-derived umbilical artery absolute velocities and their relationship to fetoplacental volume blood flow: a longitudinal study. Ultrasound Obstet Gynecol. 2005;25(5):444–453. [DOI] [PubMed] [Google Scholar]
- 33.Schabel MC, Morrell GR. Uncertainty in T(1) mapping using the variable flip angle method with two flip angles. Phys Med Biol. 2009;54(1):N1–8. [DOI] [PubMed] [Google Scholar]
- 34.Frias AE, Schabel MC, Roberts V, Tudorica A, Grigsby PL, et al. Using dynamic contrast enhanced MRI to quantitatively characterize maternal vascular organization in the primate placenta. Magn Reson Med. 2015;73(4):1570–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schabel MC, Roberts VHJ, Lo JO, Platt S, Grant KA, et al. Functional imaging of the non-human primate placenta with endogenous blood oxygen level-dependent contrast. Magn Reson Med. 2016;76(5): 1551–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khong TY, Mooney EE, Ariel I, Balmus NCM, Boyd TK, Brundler MA, et al. Sampling and definitions of placental lesions: Amsterdam Placental Workshop Group Consensus Statement. Arch Pathol Lab Med. 2016;140(7):698–713. [DOI] [PubMed] [Google Scholar]
- 37.Andrews S FastQC: A quality control tool for high throughput sequence data. 2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- 38.Ewels P, Magnusson M, Lundin S, Kaller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dobin A, , Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/btw635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeLuca DS, Levin JZ, Sivachenko A, et al. RNA-SeQc: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28(11):1530–1532. Doi. 10.1093/bioinformatics/btw196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.R Core Team (2017). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/. [Google Scholar]
- 43.Chen YA, Lun T, Smyth GK. From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Res. 2016;5:1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Law CW, Chen Y, Shi W, Smyth GK. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2): R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acid Res. 2015;43(7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Statistical Society Series B (Methodological). 1995;57(1): 289–300. [Google Scholar]
- 48.Kramer A, Green J, Pollard J, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinforma Oxf Engl. 2014;30(4):523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burd L, Roberts D, Olson M, Odendall H. Ethanol and the placenta: A review. J Matern Fetal Neonatal Med. 20(5):361–375. PMID:17674239. [DOI] [PubMed] [Google Scholar]
- 50.Myatt L Placental adaptive responses and fetal programming. The Journal of Physiology. 2006;572(Pt 1):25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davis-Anderson KL, Berger S, Lunde-Young ER, et al. Placentla proteomics reveal insights into fetal alcohol spectrum disorders. Alcohol Clin Exp Res. 2017;41(9):1551–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roberts VH, Rasanen JP, Novy MJ, et al. Restriction of placental vasculature in a non-human primate: a unique model to study placental plasticity. Placenta. 2012;33(1):73–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lo J, Schabel M, Roberts V, et al. Vitamin C supplementation ameliorates the adverse effects of nicotine on placental hemodynamics and histology in non-human primates. Am J Obstet Gynecol. 2015;212(3):370.e1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hirsch AJ, Roberts VHJ, Grigsby PL, et al. Zika virus infection in pregnant rhesus macaques causes placental dysfunction and immunopathology. Nat Commun. 2018;19(1):263. [DOI] [PMC free article] [PubMed] [Google Scholar]