Figure 1. SCN resistance of cqSCN-006 soybean roots with edited Glyma.15G191200 allele.
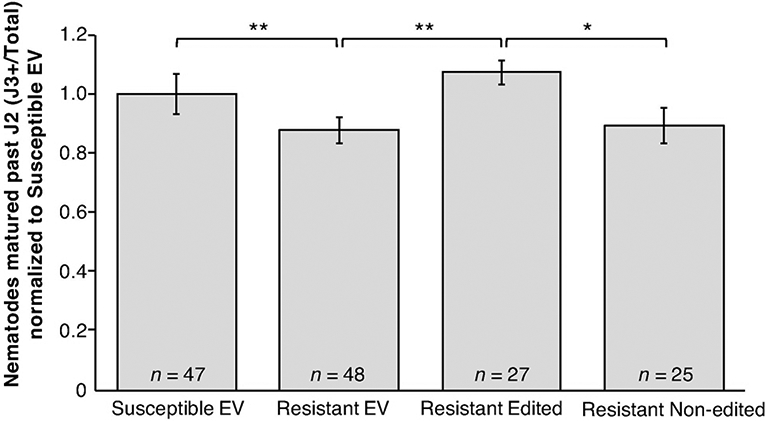
SCN development on transgenic roots of cqSCN-006 line LD10-30110 (SCN resistant) carrying CRISPR/Cas9-induced mutations in one or both alleles of Glyma.15G191200 (Resistant Edited), compared to control LD10-30092 (SCN susceptible) roots transformed with empty vector (Susceptible EV), LD10-30110 with empty vector (Resistant EV), or LD10-30110 transformed with the gene editing construct but not carrying mutations in the Glyma.15G191200 target region (Resistant Non-edited). Combined data from four independent experiments are shown; total n for each treatment is shown along with mean and standard error of the mean. Asterisks identify all significant differences (* = P < 0.1, ** = P < 0.05, *** = P < 0.01, One-tailed Welch’s t-test).
