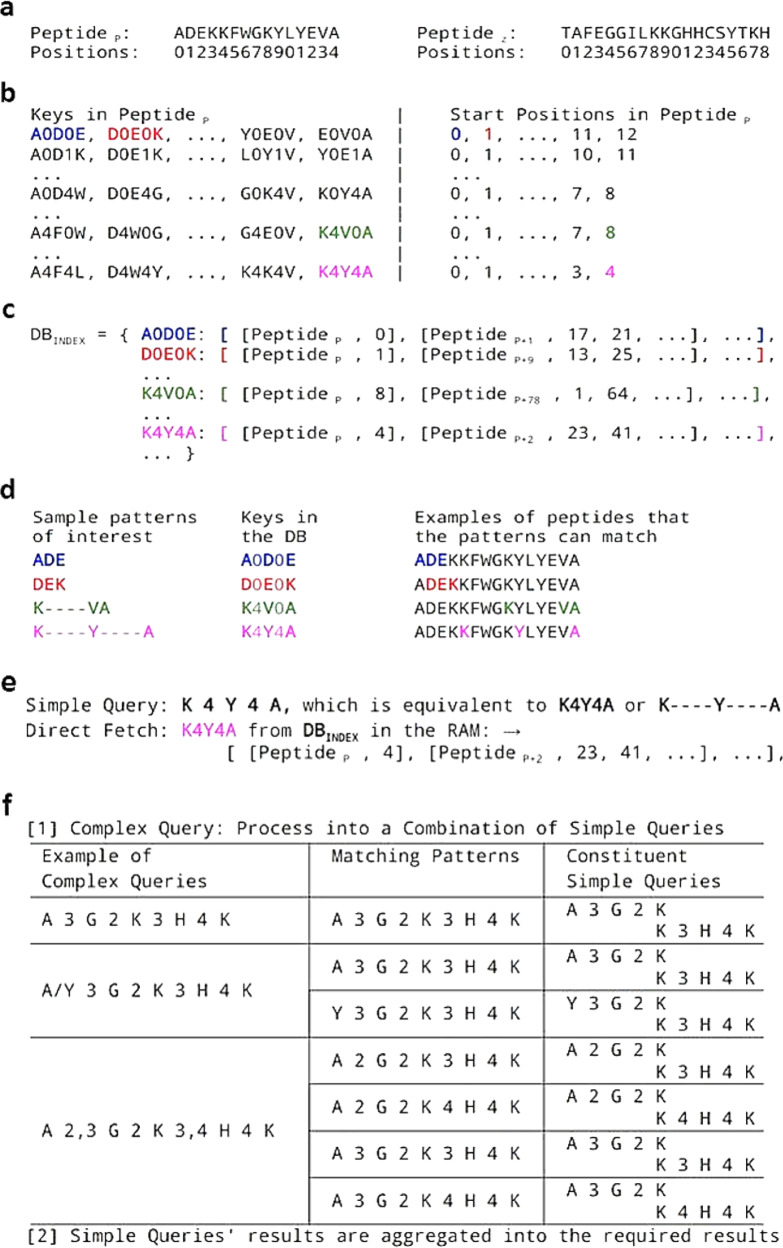
Fig. 1. Indexing the peptides and querying of TP-DB.
a The positions of amino acids in the p-th helical peptide (peptidep) are indexed from 0 to 14. b All the possible patterns that can be found in peptidep, stored as keys and values. c In TP-DB (or DBIndex), keys are paired values, and values are where these keys can be found, including in which peptides and the starting position of the keys. For instance, [A 0 D 0 E] can be found in the “0-th position” in the p-th peptide, while it can also be found in the 17th and 21st positions in the (p + 1)-th peptide. d The patterns of interest are translated into “keys” comprising amino acids’ single-letter codes separated by numbers, the sizes of the gaps. e Results for a simple query retrieved from TP-DB. f A compounded query can be broken down into subqueries and the result is a joint result from subqueries. For efficient search, two types of libraries are loaded in a timely order. What is constantly loaded in the memory, with a small memory footprint, is a collection of key–value pairs that only index which proteins have certain keys but not their locations. Only when certain proteins are visited because they contained the queried keys, then libraries of a second type, inferring the locations of keys in certain proteins, are then loaded into the memory to report the found sequences, before the protein-relevant libraries are unloaded from the memory again soon after a search job is finished. All abbreviations of amino acids are listed in Table 1.