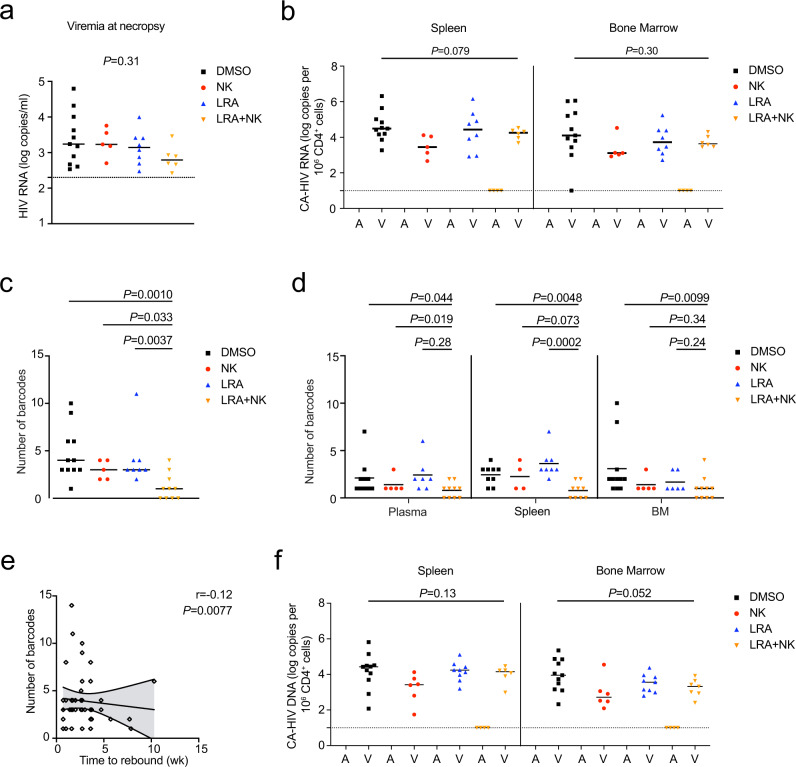
Fig. 6. Levels of viral RNA, barcode diversity, and HIV DNA from infected mice treated with SUW133 and NK cells.
a Plasma viral loads of rebounding mice. n = 11 biologically independent animals in DMSO (black) group, n = 5 biologically independent animals in NK only (red) group, n = 8 biologically independent animals in LRA only (blue) group, n = 6 biologically independent animals in LRA plus NK (orange) group. b Cell-associated “CA” HIV RNA from the spleen and bone marrow “BM” of mice that were aviremic “A” or had rebound viremia “V” at necropsy. c, d Number of unique barcodes quantified by deep sequencing of viral RNA from pooled organs per mouse (c) or from the plasma, spleen, and BM of each mouse (d). e Scatterplot of a number of unique barcodes per mouse and time to rebound among all mice that rebounded after ART interruption. n = 48 biologically independent rebounding animals. Lines are linear predictions of time to rebound on the number of barcodes. The 95% confidence intervals of the fitted values are shown by gray areas. r Pearson correlation coefficient. f Total CA-HIV DNA from the spleen and BM of mice that were aviremic or had rebound viremia at necropsy. n = 11 biologically independent animals in DMSO only group, n = 5 biologically independent animals in NK only group, n = 8 biologically independent animals in LRA only group, n = 10 biologically independent animals in LRA plus NK group (b–d, f). Black dotted line indicates the detection limit of 2.3 log RNA copies per ml (a) or 1.0 log RNA or DNA copies (b, f). Horizontal bars represent the means (a–d, f). P values were calculated using one-way ANOVA Kruskal–Wallis test (a, b, f), two-tailed Mann–Whitney (c, d), or Pearson correlation test (e). Results are pooled from two independent experiments. Source data are provided as a Source Data file.