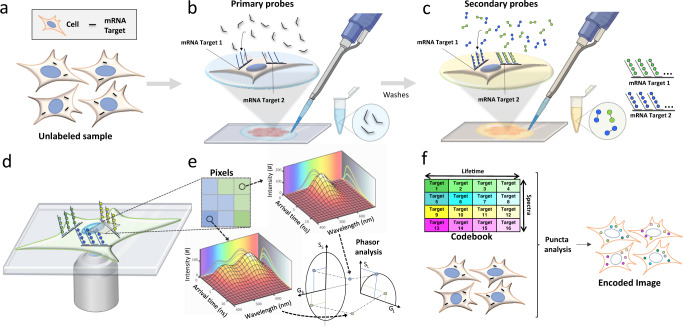
Fig. 1. Schematic of the MOSAICA approach for labeling and analysis of spectral and time-resolved components.
a Sample(s) can be fixed cells or tissues. RNA transcripts from genes of interest are targeted for detection. Protein targets can be stained too in mRNA and protein codetection. b Primary labeling probes are designed to include two functional regions: a target region which is complementary and can bind to the mRNA target and an adjacent readout region which can subsequently bind to fluorescently labeled oligonucleotides. c Secondary fluorescent probes are added to bind to the primary probes to form different combinations (combinatorial labeling) through a “readout” domain. d Labeled targets are measured under a fluorescent microscope to interrogate the spectral and lifetime characteristics of the labeled moieties. e Phasor analysis is used to identify which fluorophore labels are present in each pixel and puncta. f Labeled targets eliciting the encoded intensity-based and time-based signature are decoded to reveal the locations, identities, counts, and distributions of the present mRNA targets in a multiplexed fashion.