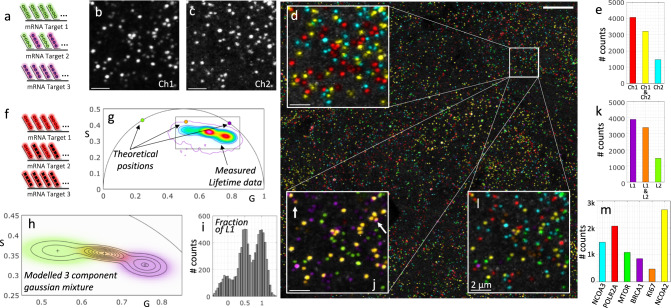
Fig. 3. Working example of combinatorial labelling of three mRNA targets with two probes.
a Transcripts of three different target genes are tagged using two probes with different spectra. Targets 1 and 3 are tagged each with one probe, Target 2 is tagged with both simultaneously. b, c The fluorescence is collected in the two expected spectral channels for the known emission of the two probes (representative small regions of a whole 3D field of view). d The maximum projection of the two channels is shown and pseudo-colored depending on the presence in the respective channels (as an inset within the whole field of view. e The actual counts of each target within the whole field of view. f As a parallel example, transcripts of three different target genes are tagged using two probes with different lifetime. Targets 1 and 3 are tagged each with one probe, Target 2 is tagged with both simultaneously. g The phasor plot presents three populations, corresponding to the pixels with the three combinations; the two components by themselves plus the linear combination falling in the middle. h) Machine learning clustering technique is used to identify the groups (Gaussian mixture model). i) The multicomponent method is used to extract the fraction of one of the components in each detected puncta. j The same inset is shown with the pseudocoloring now depending on the lifetime clustering. k The counts for each lifetime cluster in the whole field of view. l The combination of the information in both the spectral and the lifetime dimension yields a final 6-plex. m The overall counts for the 6-plex detection of transcripts including POLR2A (Alexa647 & ATTO565), MTOR (ATTO647 & ATTO565), KI67 (Alexa647 & ATTO647), BRCA1 (Alexa647), NCOA2 (ATTO647), NCOA3 (ATTO565) with the appropriate expressed genes that correspond to each combination. Experiments were conducted with cultures of mNeon green cells. Scale bar 10 µm in large image and 2 µm in insets. Source data are provided as a Source Data file.