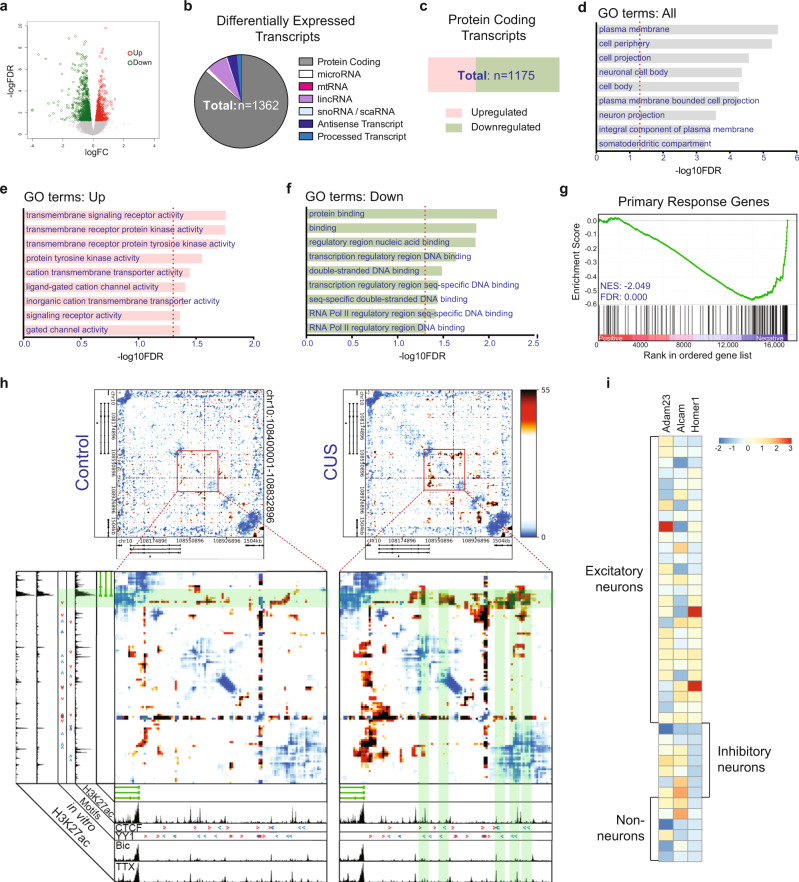
Fig. 2. CUS impacts transcription and chromatin folding in the PFC.
a Significantly upregulated transcripts (red) and downregulated transcripts (green) from comparison of control and CUS PFC nuclear transcripts (FDR < 0.05). b Piechart exhibiting representation of differentially expressed RNA transcript populations. c Barchart portraying makeup of upregulated and downregulated differentially expressed protein-coding transcripts. d–f Top 9 most significantly enriched gene ontology (GO) terms for d all, e upregulated, and f downregulated CUS DEGs (FDR < 0.05). g GSEA plot charting negative enrichment of neuronal primary response genes in list of differentially expressed genes obtained from bulk nuclear RNA-seq analysis of control and CUS PFCs. NES, normalized enrichment score. h Background-corrected interaction frequency heatmaps displaying chromatin contacts in a 1.5-Mb region surrounding the Syt1 gene in control and CUS frontal cortical tissues. The highlighted region in each heatmap marks the location of a zoomed-in plot. H3K27ac ChIP-seq track from adult cortical excitatory neurons control is shown below each heatmap. Enlarged heatmaps plot genomic locations and directionality of CTCF and YY1 binding sequences (red and blue arrowheads) in regions showing increased chromatin interactions in CUS mice (green shaded boxes). Legend depicts background-corrected interaction scores. i Heatmap showing expression of CUS DEGs, Homer1, Adam23, and Alcam, across the cortical cell populations defined by single-nucleus RNA-seq in the adult mouse neocortex. Legend depicts z-score of normalized gene expression.