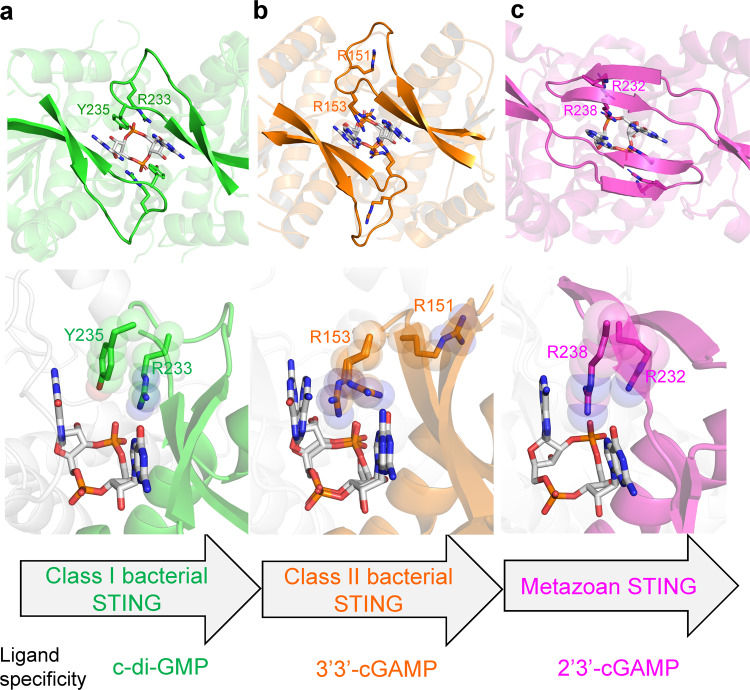
Fig. 7. Structural comparison of bacterial and metazoan STING suggests a model for the evolution from c-di-GMP based signaling in prokaryotes to the 2’3’-cGAMP-dependent signaling in eukaryotes.
The enlarged view of ligand-binding site and β-strand lid region of (a) PcSTING-c-di-GMP (green), (b) FsSTING-3’3’-cGAMP (orange, PDB: 6WT4), and (c) human STING-2’3’-cGAMP complexes (magenta, PDB: 4KSY). The cyclic dinucleotide ligands are shown as white sticks. The class I bacterial STING has conserved RX(Y/F) motif (R233-X-Y235 in PcSTING) in lid region and specifically recognizes c-di-GMP by four-layer stacking interaction. The class II bacterial STING has conserved RXR motif in the β-strand lid (R151-X-R153 in FsSTING), but recognizes 3’3’-cGAMP asymmetrically. The conserved arginine residues of metazoan STING mainly recognize the 2’-5’/3’-5’ phosphodiester linkage of 2’3’-cGAMP instead of the nucleobases. The class II bacterial STING is more closely related to the metazoan STING and is proposed to be acquired into the ancestor of early metazoan.