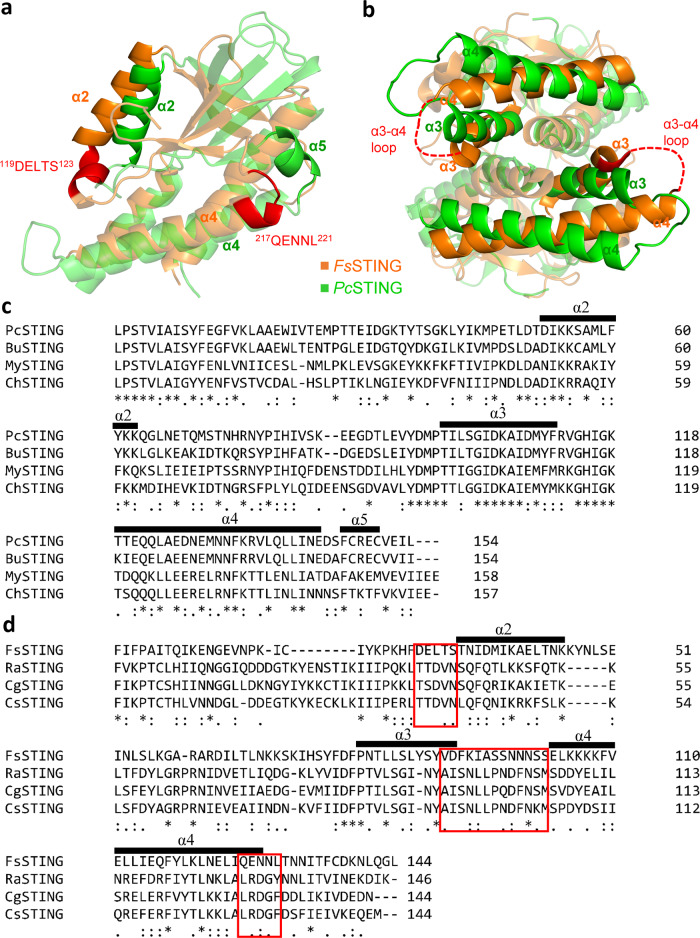
Fig. 8. The differences in oligomerization interface between Class I and Class II bacterial STING.
a The predicted oligomerization interface at the beginning of α2 helix and the end of α4 helix of FsSTING (orange, PDB: 6WT4) are indicated and colored in red. By contrast, PcSTING (green) protomer uses the α5 helix to interact with the whole α2 helix of the adjacent PcSTING protomer. b The PcSTING and FsSTING dimers are superimposed. The orientation of α3 and α4 helix and thereby the predicted α3-α4 loop of FsSTING (red dashed lines) are different from those of PcSTING. c Multiple sequence alignment of four representatives of Class I bacterial STING proteins. d Multiple sequence alignment of four representatives of Class II bacterial STING proteins. The predicted oligomerization interfaces in FsSTING are highlighted by red frames.