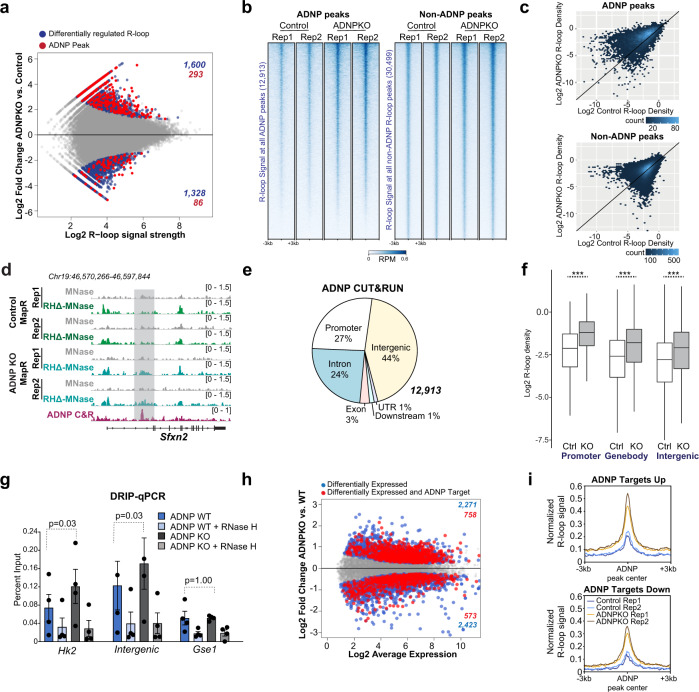
Fig. 5. ADNP suppresses R-loops at its binding sites in vivo.
a MA plot of MapR signal in 61,652 R-loop peaks between Control (ADNP-KI) and ADNP KO mESCs. Blue dots indicate 2928 significant differential R-loop sites (FDR < = 0.05, DiffBind; 1600 up and 1328 down in KO). Significant differential R-loops that overlap an ADNP CUT&RUN peak (293 up, 86 down) are colored in red. b Heatmap of normalized MapR signal across 6-kb windows centered on ADNP peak locations (left) or control R-loop peaks that do not overlap ADNP peaks (right). Rows are sorted in decreasing order by mean signal across all samples. I Hexagonal-binned scatterplot of mean normalized MapR signal in control and ADNP KO mESCs across the regions shown in (b). d Genome browser view of the Sfxn2 gene showing MapR signal (RPM) in control and ADNP KO mESCs and ADNP CUT&RUN signal (RPM) in control mESCI. e Pie chart displaying distribution of 12,913 ADNP peaks across genomic features. f Boxplot displaying log2 normalized R-loop read densities in control and ADNP KO mESCs across ADNP peaks, grouped by genomic feature. ***p < 2.2 × 10−16 (Welch’s two-sided t-test; n = 3432 promoter peaks, 3653 gene body peaks, or 5828 intergenic peaks). Box, 25th percentile – median – 75th percentile. Whiskers extend to 1.5x interquartile range; outliers not displayed. g DRIP-qPCR results using the S9.6 antibody in WT and ADNP KO mESCs at an Hk2 gene region, an intergenic region, and a Gse1 gene control region. Bar chart, mean ± SEM; individual values shown as Dots. p, Welch’s two-sided t-test (n = 4 biologically independent samples). h MA plot of RNA-Seq expression for 16,195 genes between WT and ADNP KO mESCs. Blue dots indicate differentially expressed genes (adjusted p-value <=0.05; p-values computed by edgeR with Benjamini–Hochberg adjustment for multiple comparisons). Red dots indicate differentially expressed genes with an ADNP CUT&RUN peak within 3 kb of the gene body. i Signal plot of normalized control and ADNP KO MapR signal over ADNP peaks associated with ADNP target genes that are upregulated (1219 peaks) or downregulated (1040 peaks) in ADNP KO mESCs relative to WT. Source data underlying (g) are provided as a Source data file.