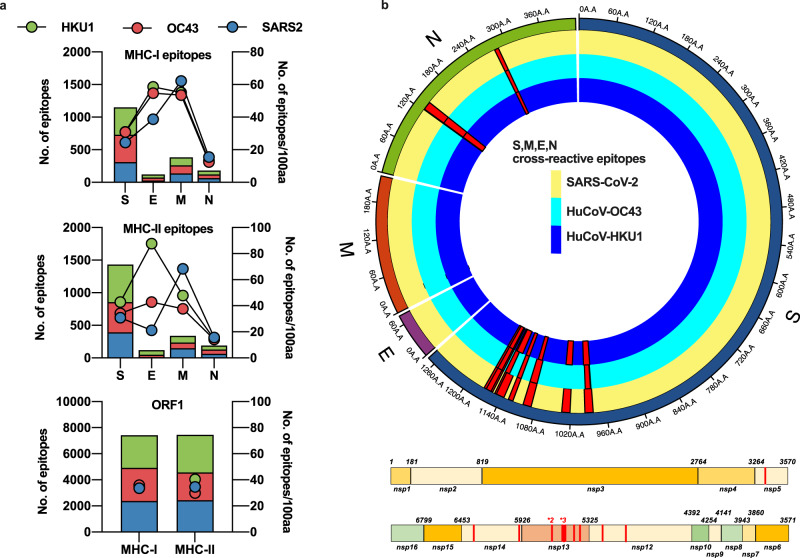
Fig. 1. In silico prediction of putative epitopes in human endemic beta-coronaviruses and SARS-CoV-2.
Available sequences for S, M, E, N and ORF1 from SARS-CoV-2, huCoV-OC43 and huCoV-HKU1 were assessed for potential HLA-binding motifs by SYFPEITHI and IEDB as described in the Methods. The number of predicted epitopes is presented in the stacked bar charts and the immunogenicity for each protein of each virus is depicted as points on the right Y-axis. Green bars and circles depict huCoV-HKU1 (HKU1), red huCoV-OC43 (OC43) and blue SARS-CoV-2 (SARS2) (a). The predicted cross-reactive epitopes and their prevalence within each virus for all sequences available are mapped as red histograms in a linear plot for ORF1 and a radial plot for S, M, E and N. All mapped cross-reactive epitopes within ORF1 were present across SARS-CoV-2, HKU1 and OC43. Within the radial plot, the yellow track represents SARS-CoV-2 sequence, dark blue HKU1 and light blue OC43, with red histograms representing the prevalence of the putative epitope across the viruses (b).