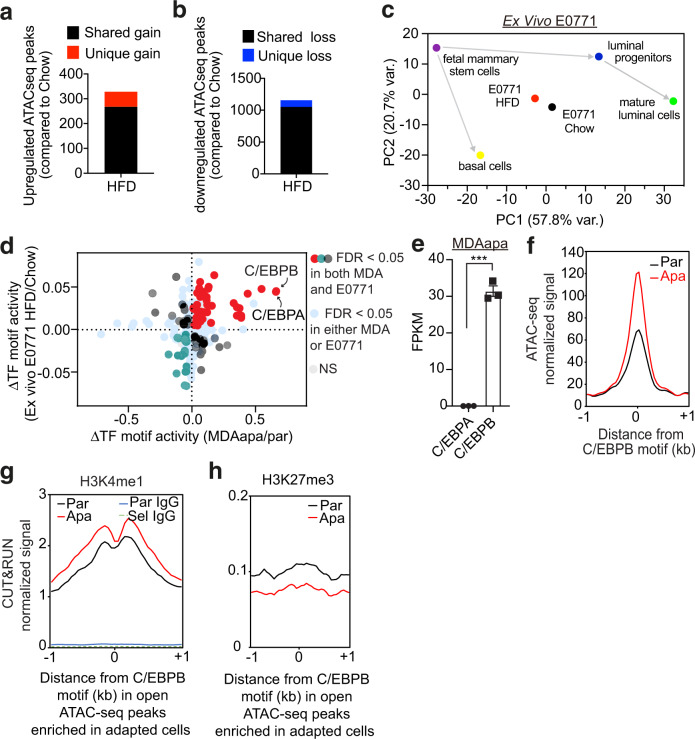
Fig. 3. Adaptation to obese environments induces open chromatin linked with C/EBPB occupancy.
a, b Total number of significantly upregulated (a) and downregulated (b) ATACseq peaks in E0771 HFD (n = 3) relative to chow (n = 4) by DiffBind with a false discovery rate (FDR) < 0.05. Unique gain (a) or loss (b) peaks refer to the peaks identified only in the HFD or chow condition, respectively, whereas shared peaks are peaks called in both conditions. c Principal component analysis showing principal components (PC) 1 and 2 of E0771 ex vivo cells and different cell lineages along the mammary gland developmental trajectory (GEO: GSE116386) using the average transcription factor motif activity estimated by chromVar. d Overlap of differential transcription factor binding motif activity between MDA-MD-231 (apa/par) and E0771 (HFD/Chow) as determined by diffTF. e Within-sample normalized gene expression of transcription factor homologs C/EBPA and C/EBPB in PA-adapted MDA-MB-231 cells using RNA-seq. FPKM = fragments per kilobase of transcript per million mapped reads. Data are represented as mean ± SEM of 3 replicates from each group. Statistical significance determined with unpaired, two-tailed Student’s t-test (P < 0.0001). f Metagene representation of the mean ATACseq signal across more accessible C/EBPB motif regions in parental (n = 3) or adapted (n = 3) MDA-MB-231 cells. The mean signal of three adapted or parental MDA-MB-231 biological replicates was determined by averaging signals of 1 kb around the center of C/EBPB DNA-binding motifs. g, h Metagene representation of the mean H3K4me1 (par n = 2; apa n = 2) and IgG signals (g) and the mean H3K27me3 signals (par n = 3; apa n = 3) (h) across more accessible C/EBPB motif regions as in (f) in MDA-MB-231 cells. The mean signals of biological replicates were determined by averaging signals of 1 kb around the center of C/EBPB DNA-binding motifs. Source data are provided as a Source data file.