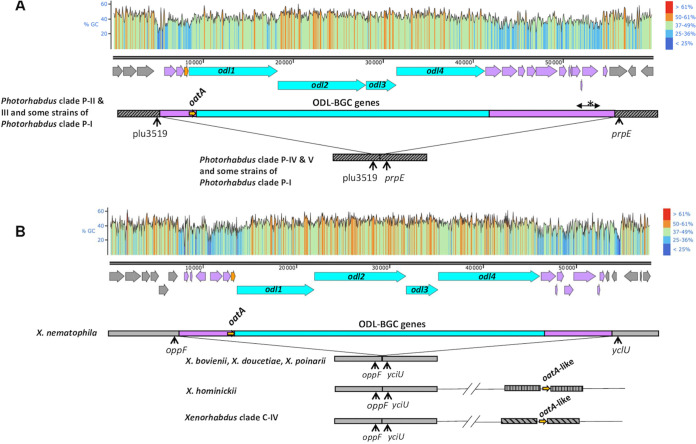
FIG 6.
Genomic environment of the oatA gene in the Photorhabdus (A) and Xenorhabdus (B) genomes. Blue box, odl-BGC region; pink boxes, genomic regions framing the odl-BGC region encoding several enzymes, including OatA (the asterisk shows a highly shuffled subregion with a variable gene content according to the strain); gray boxes, core genome (different motifs indicate different regions of core genomes). GC content ({G or C}/{A, T, G, or C}) was calculated for a sliding window with a length of 25 nucleotides and plotted with SnapGene software (from Insightful Science). (A) Strains from clade P-I containing the oatA ortholog include P. luminescens subsp. luminescens DSM 3368, P. luminescens HIM3, P. namnanoensis PB45.5, and Photorhabdus sp. H1. Strains from the clade P-I which totally lack the oatA ortholog include P. aegyptiae BA1, Photorhabdus sp. LN2, NBAII HiPL 101, and NBAII Hb 105. Strains from clade P-II include P. bodei CN4 and LJ24-63, P. kayaii M-HU2, C-HU2, and HUG-39, P. kleinii S9-53 and S10-54, and Photorhabdus sp. CRCIAP01. Strains from clade P-III include P. laumondii TT01, HP88, and IL9. Strains from clade P-IV include P. asymbiotica ATCC 43949; P. australis subsp. australis DSM 17609, P. cinnerea DSM 19724, and P. heterorhabditis VMG. Strains from clade P-V include P. temperata J3, Meg1, and M1021, P. thracensis DSM 15199, P. khani NC19, and P. tasmaniensis DSM 22387. (B) X. nematophila strains include F1, ATCC 19061, K102 (CNCMI-4530), Anatoliense, Websteri, and C2-3. X. hominickii strains include ANU1 and DSM 17903. Strains from clade C-IV include Xenorhabdus sp. KK7.4 and KJ12.1, X. innexi HGB1681 and DSM 16336, X. stockiae DSM 17904, and X. cabanillassii 17905 and JM26. The oatA ortholog is totally absent from the strains of Xenorhabdus phylogenetic clades C-I and C-III, illustrated here by the strains X. bovienii CS03, X. doucetiae FRM16, and X. poinarii G6.