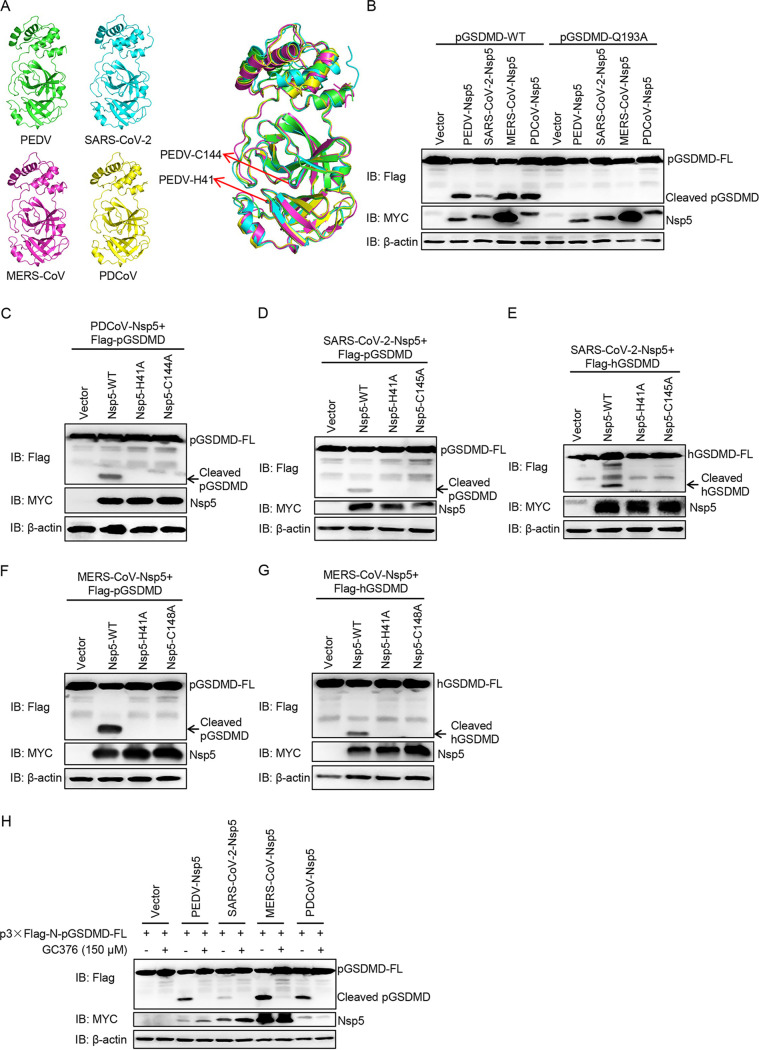
FIG 7.
GSDMD is a common substrate of different coronaviruses Nsp5. (A) Structure alignment of CoVs Nsp5. Red arrows indicate conserved enzymatic proteolysis residues His41 and Cys144. The three-dimensional structures were derived from the Protein Data Bank with the following accession numbers: PEDV, 4XFQ; SARS-CoV-2, 7BUY; MERS-CoV, 5WKK; PDCoV, 6JIJ. (B) HEK293T cells were cotransfected with plasmids encoding p3×Flag-N-pGSDMD-FL or p3×Flag-N-pGSDMD-FL-Q193A and Nsp5 encoded by PEDV, PDCoV, SARS-CoV-2 and MERS-CoV. After 24 h, cells were lysed and detected by immunoblotting. (C) HEK293T cells were cotransfected with plasmids encoding pGSDMD and wild-type PDCoV Nsp5 or its protease-defective mutants (H41A and C144A). After 24 h, cells were lysed for immunoblotting. (D and E) HEK293T cells were cotransfected with plasmids encoding pGSDMD and wild-type SARS-CoV-2 Nsp5 or its protease-defective mutants (H41A and C145A), hGSDMD, and wild-type SARS-CoV-2 Nsp5 or its protease-defective mutants (H41A and C145A). After 24 h, cells were lysed for immunoblotting. (F and G) HEK293T cells were cotransfected with plasmids encoding pGSDMD and wild-type MERS-CoV Nsp5 or its protease-defective mutants (H41A and C148A), hGSDMD, and wild-type MERS-CoV Nsp5 or its protease-defective mutants (H41A and C148A). After 24 h, cells were lysed for immunoblotting. (H) HEK293T cells were cotransfected with plasmids encoding p3×Flag-N-pGSDMD-FL and Nsp5 encoded by PEDV, PDCoV, SARS-CoV-2, and MERS-CoV. At 6 h after transfection, cells were mock treated or treated with GC376 (final concentration of 150 μM). After 24 h, cells were lysed for immunoblotting.