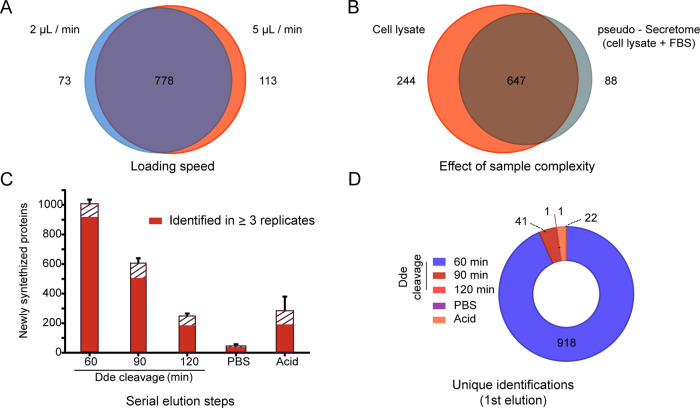
Figure 2.
Optimization of the automated protocol. (A) Comparison of two different loading speeds based on the number of identified NSPs after loading three sample replicates. Sample is a cell lysate. (B) Effect of increasing sample complexity, achieved by the addition of FBS to the cell lysate, to imitate a secretome sample obtained in complete cell culture media. Newly synthesized proteins from three sample replicates loaded at 5 μL/min are shown. (C) Elution of captured proteins by either chemical cleavage of the Dde group present in the conjugate through incubation with 2% hydrazine, PBS wash, or denaturing biotin with 5% acetic acid. Sample is a cell lysate. The error bar represents the standard error of the mean of four replicates. The solid red filling represents NSPs enriched consistently in three or more replicates of the same condition. (D) NSPs obtained in C organized to showcase the first elution step in which they eluted consistently in three replicates.