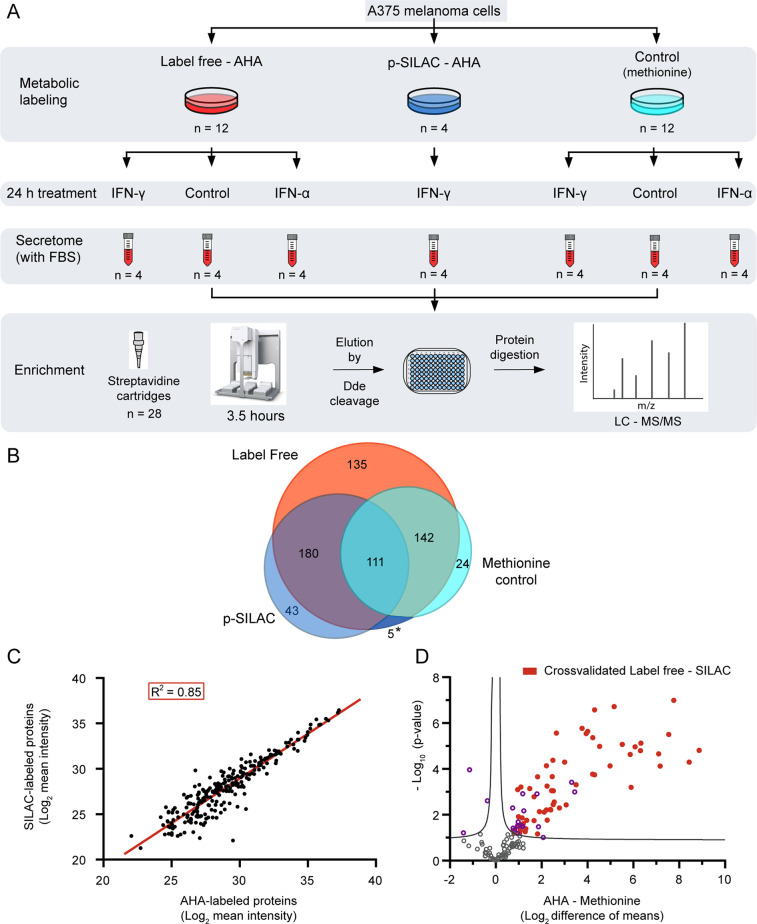
Figure 3.
Performance of the automated method in a label-free approach to study melanoma secretory response to IFN treatment. (A) Schematic representation of the workflow for label-free identification of NSPs. The plates were seeded, treated, and collected simultaneously in one experiment and secretomes were enriched with the workflow shown in Figure 1B. The control in the metabolic labeling section consists of the substitution of AHA with methionine in the culture media. The controls in the 24 h treatment section consist of untreated samples. The secretomes correspond to the collection of the complete media (including FBS) after the treatment, without detachment of the cells. (B) Proteins obtained by label-free identification in the presence or absence of AHA are compared to the p-SILAC proteins filtered by the presence of the label. The asterisk (*) area is shared between p-SILAC and methionine control. (C) Linear correlation between 240 mutual proteins obtained after AHA enrichment by the label-free and p-SILAC approaches. Proteins shown have at least two values per condition. A linear fit has been applied (red line) with an R2 = 0.85 and a slope of 0.98. (D) Statistical comparison of 144 label-free proteins identified in both the AHA-labeled proteins and methionine controls (unspecific binding), in at least three replicates. Proteins above the curve are significantly over-represented in the presence of AHA (right) or methionine (left). Proteins in red are cross-validated as newly synthesized by the results in the p-SILAC dataset. Curve represents FDR = 0.05.