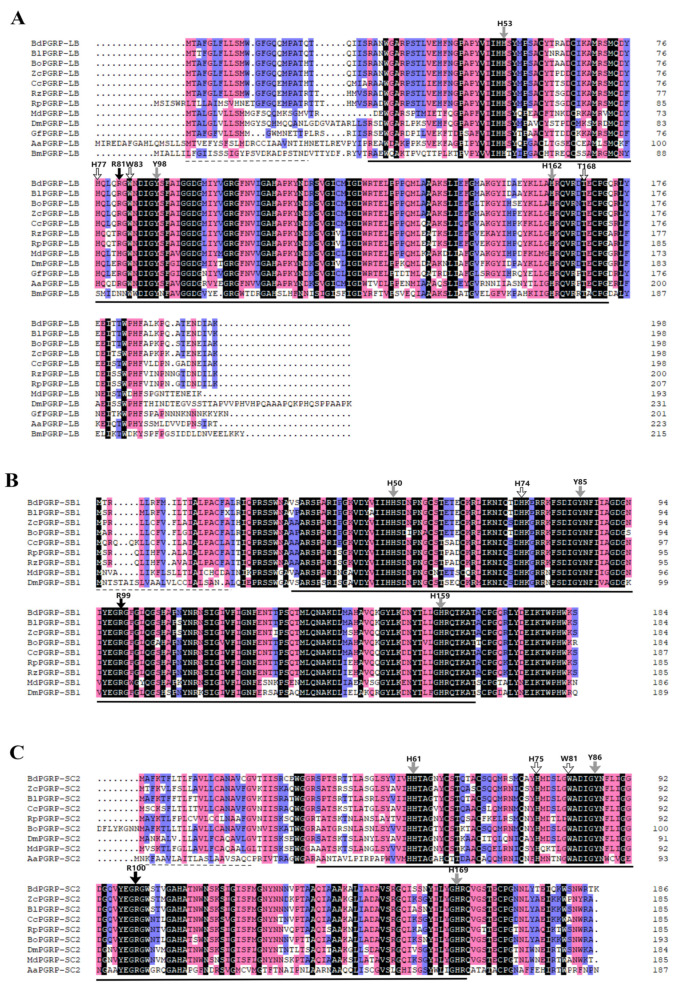
Figure 1.
Amino acid sequence alignment of BdPGRPs with that of homologous genes in other insect species. (A) Multiple alignments of PGRP-LB. BdPGRP-LB was aligned with Bactrocera latifrons PGRP-LB (XP_018789449.1), Bactrocera oleae PGRP-LB (XP_014091181.2), Zeugodacus cucurbitae PGRP-LB (XP_011197144.1), Ceratitis capitate PGRP-LB (XP_004518089.1), Rhagoletis zephyria PGRP-LB (XP_017470705.1), Rhagoletis pomonella PGRP-LB (XP_036322481.1), Aedes aegypti PGRP-LB (XP_021709443.1), Drosophila melanogaster PGRP-LB (NP_731575.1), Bombyx mori PGRP-LB (XP_012548100.1), Musca domestica PGRP-LB (XP_005180889.1), and Glossina fuscipes PGRP-LB (ACI22620.1). (B) Multiple alignments of PGRP-SB. BdPGRP-SB1 was aligned with B. latifrons PGRP-SB (XP_018789286.1), B. oleae PGRP-SB (XP_014099773.1), Z. cucurbitae PGRP-SB (XP_011181375.1), C. capitate PGRP-SB (XP_004537949.1), R. zephyria PGRP-SB (XP_017486043.1), R. pomonella PGRP-SB (XP_036336342.1), D. melanogaster PGRP-SB (CAD89135.1), M. domestica PGRP-SB (NP_001295929.1), and B. mori PGRP-SB (XP_004929843.1). (C) Multiple alignments of PGRP-SC2. BdPGRP-SC2 was aligned with B. latifrons PGRP-SC2 (XP_018798904.1), B. oleae PGRP-SC2 (XP_014085196.2), C. capitate PGRP-SC2 (XP_004520319.1), Z. cucurbitae PGRP-SC2 (XP_011180165.1), R. pomonella PGRP-SC2 (XP_036334551.1), M. domestica PGRP-SC2 (XP_005184140.3), D. melanogaster PGRP-SC2 (CAD89184.1), A. aegypti PGRP-SC2 (XP_011492940.1), and B. mori PGRP-SC2 (XP_004929814.1). The identical amino acids are shown against a black background; 75% conserved amino acids are shown against a pink background; 50% conserved amino acids are shown against a blue background. The signal peptides are indicated by dashed lines. The amidase domains are indicated by solid lines. Black arrows indicate the amino acid residues required for the recognition of DAP-type peptidoglycan. Grey arrows indicate the amino acid residues required for Zn2+ binding. White arrows indicate the amino acid residues required for amidase activity.