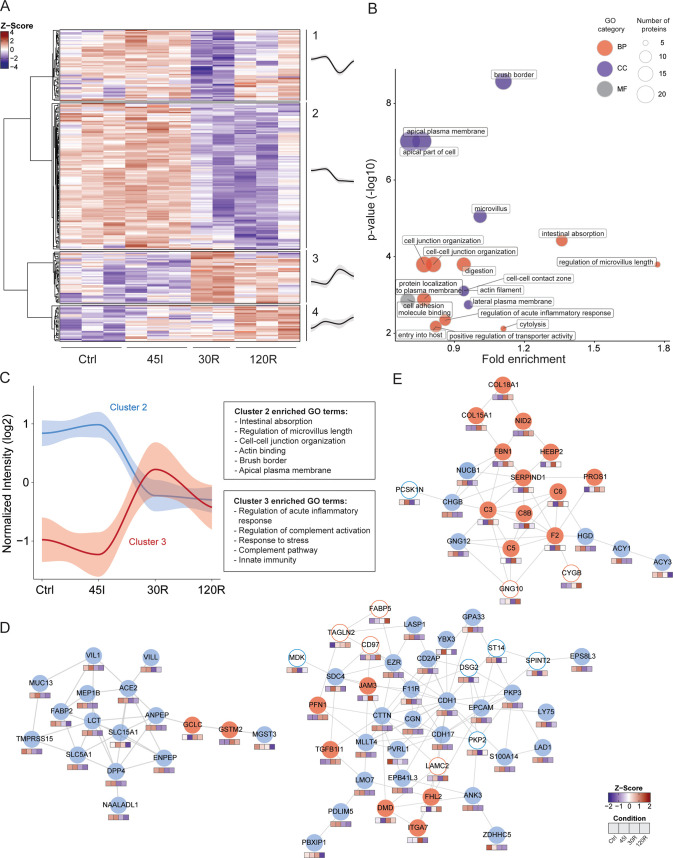
Figure 2.
Dynamic proteome during ischemia–reperfusion in the human intestine. (A) Heatmap visualizing clustering of differentially expressed proteins. Hierarchical clustering was based on the Z scores of the log 2 values of differentially expressed proteins. The ANOVA test was performed and P < 0.05 was considered statistically significant. Average temporal profiles are shown for every cluster (a gray area represents CI). (B) Functional enrichment analysis of differentially expressed proteins. Over-represented gene ontology (GO) terms are shown (P < 0.05). GO term fold enrichment (log 10[observed/expected]) is plotted against P-value (−log 10). The size of the dot correlates with the number of proteins linked to that GO term, as indicated in the legend. Red dot, GO Biological Processes; blue dot, GO cellular component, gray dot, GO molecular function. (C) Dynamic profile for proteins in clusters 2 (blue) and 3 (red) is shown, as well as corresponding overrepresented GO terms. (D, E) Protein networks showing interacting proteins. Networks were generated using STRING. The color of the circle indicates the temporal profile (cluster) of the protein. Blue line, cluster 1; blue fill, cluster 2; red fill, cluster 3, red line, cluster 4. Color cubes below each protein indicate the Z-score intensity in Ctrl, 45I, 30R, and 120R, respectively. The majority of proteins in the networks in (D) are located in cluster 2 (blue fill). The majority of proteins in the network in (E) are located in cluster 3 (red fill).