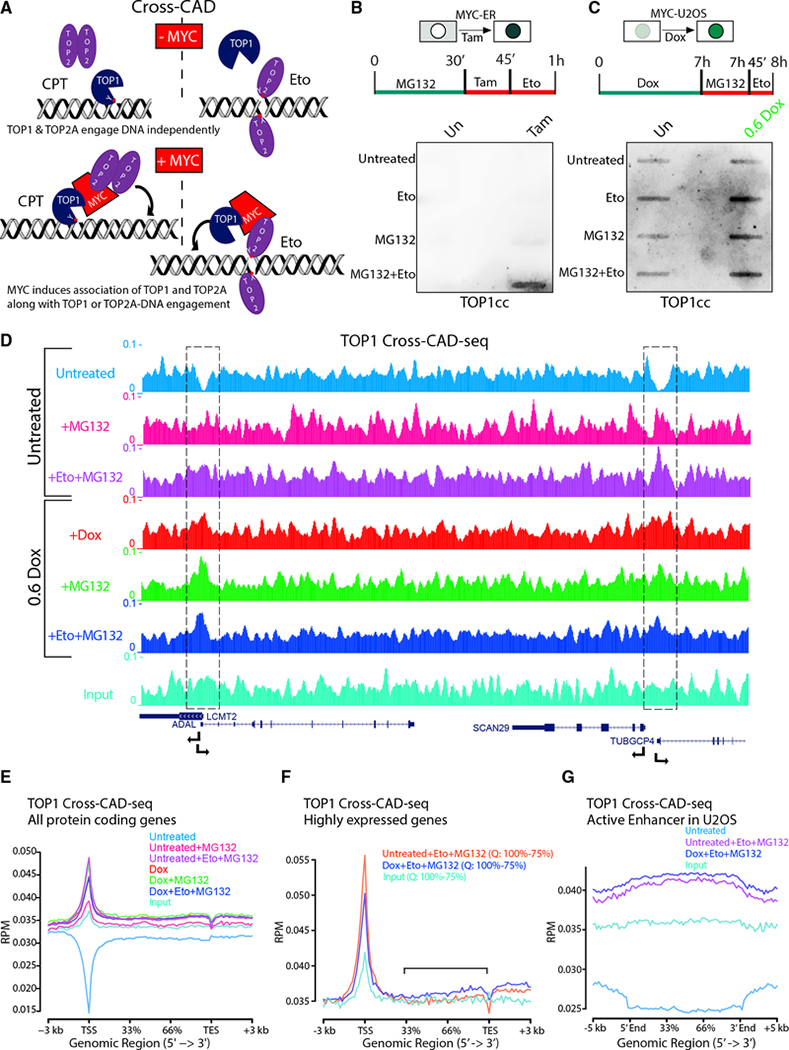
Figure 6. The topoisome engages promoters and enhancers to favor transcription.
(A) Scheme of “cross-CAD” assay. CPT increased engagement of TOP2A as well as TOP1 in the presence of MYC, while Eto increased engagement of TOP1 as well as TOP2A. Red dots with Y indicate the active site of TOP1 (Y723) or TOP2 (Y805).
(B) Detection of MYC-tethered TOP1 along with Eto-trapped TOP2Acc by cross-CAD assays in HO15.19-MYC-ER cells with or without Tam with anti-TOP1.
(C) As in (B) but in Dox-induced U2OS-MYC-EGFP cells.
(D) Genome Browser of TOP1 cross-CAD-seq (RPM) showing two pairs of divergent promoters (dashed boxes).
(E and F) Average TOP1 cross-CAD-seq (RPM) at all RNAPII genes (between TSS and transcription end site [TES]) (E) or at highly (75%–100%; Q, quartile) expressed genes (F) and at active enhancers (G) in U2OS cells.