Abstract
Simple Summary
Several statistical models exist to predict a person’s risk of breast cancer. Risk assessment models can guide cancer screening approaches by identifying individuals who would benefit from additional screening. In this study, we compared the performance of four models in predicting the 5-year risk of breast cancer in a cohort of women aged 40–84 years who underwent screening mammography at three large health systems. Models showed comparable discrimination (ability to distinguish between cases and non-cases) and calibration (ability to accurately predict risk) overall, with no difference by race. Model discrimination was poorer for some cancer subtypes, and better for women with high BMI. The combined BRCAPRO+BCRAT model had improved calibration and discrimination among women with a family history of breast cancer. Our results can inform risk-based screening approaches by identifying women at a high risk of breast cancer.
Abstract
(1) Background: The purpose of this study is to compare the performance of four breast cancer risk prediction models by race, molecular subtype, family history of breast cancer, age, and BMI. (2) Methods: Using a cohort of women aged 40–84 without prior history of breast cancer who underwent screening mammography from 2006 to 2015, we generated breast cancer risk estimates using the Breast Cancer Risk Assessment tool (BCRAT), BRCAPRO, Breast Cancer Surveillance Consortium (BCSC) and combined BRCAPRO+BCRAT models. Model calibration and discrimination were compared using observed-to-expected ratios (O/E) and the area under the receiver operator curve (AUC) among patients with at least five years of follow-up. (3) Results: We observed comparable discrimination and calibration across models. There was no significant difference in model performance between Black and White women. Model discrimination was poorer for HER2+ and triple-negative subtypes compared with ER/PR+HER2−. The BRCAPRO+BCRAT model displayed improved calibration and discrimination compared to BRCAPRO among women with a family history of breast cancer. Across models, discriminatory accuracy was greater among obese than non-obese women. When defining high risk as a 5-year risk of 1.67% or greater, models demonstrated discordance in 2.9% to 19.7% of patients. (4) Conclusions: Our results can inform the implementation of risk assessment and risk-based screening among women undergoing screening mammography.
Keywords: breast cancer, risk prediction, mammography
1. Introduction
While breast cancer mortality has fallen over the past decade in the U.S., it remains the second leading cause of cancer death among women, with 43,600 breast cancer deaths projected in 2021 [1]. Identification of patients at high risk of developing breast cancer could allow targeting of preventive and screening interventions to mitigate risk to further reduce mortality. Specifically, risk-based screening approaches that tailor screening initiation, interval, and supplemental screening to individual risk may increase benefits and reduce harms of screening. Multiple validated risk assessment models have been developed to quantify an individual woman’s risk of developing breast cancer [2]. The Breast Cancer Risk Assessment Tool (BCRAT, also known as the Gail model) utilizes age, race/ethnicity, history of breast biopsy and atypical hyperplasia, first-degree family history of breast cancer, age at menarche, and age at first birth to estimate risk [3,4,5,6]. The Breast Cancer Surveillance Consortium (BCSC) model [7] utilizes age, race/ethnicity, first-degree family history of breast cancer, breast biopsy and benign breast disease, and breast density, as measured by the American College of Radiology Breast Imaging Reporting and Database System [8]. The BRCAPRO model [9] was developed in the setting of women undergoing genetic counseling and uses a detailed family history of breast and other cancers to estimate risk both of a BRCA1/2 mutation and risk of breast cancer. Recently, the BRCAPRO model was combined with the BCRAT model to create the BRCAPRO+BCRAT model [10] that incorporates both factors in the BCRAT model and detailed family history.
Our prior work compared the BCRAT, BCSC, and BRCAPRO models in a cohort of women undergoing mammography screening at a single institution [11]. We found comparable moderate discrimination and good calibration of these three models; however, we were unable to assess differences in model performance by race/ethnicity due to the lack of diversity in the cohort. Few studies have evaluated the performance of breast cancer risk models by race/ethnicity. Additionally, while we found poorer predictive accuracy of the models for HER2+ and triple-negative breast cancers, the numbers of cancers of these subtypes were small, limiting our ability to draw strong conclusions.
The purpose of the current study is to compare the performance of breast cancer risk models for use in the setting of mammography screening, and specifically to evaluate model performance among subgroups defined by race/ethnicity, by molecular subtypes, by family history, and by obesity. Additionally, we performed validation of the recently developed BRCAPRO+BCRAT model [10], utilizing three large mammography screening cohorts. We evaluated the performance of BCRAT, BCSC, BRCAPRO, and BRCAPRO+BCRAT models because these models utilize risk factors that are routinely collected during the course of clinical care in these three health systems, and therefore the integration of one or more of these risk models into clinical care for decision making is potentially feasible.
2. Materials and Methods
2.1. Study Population
We assembled three cohorts of women presenting for screening mammography at Massachusetts General Hospital (MGH), Newton-Wellesley Hospital (NWH), and the University of Pennsylvania Health System (UPenn) between 2006–2015, 2006–2015, and 2011–2015, respectively. All mammograms included were digital or digital breast tomosynthesis... The following patient-reported risk factors were collected via questionnaire at the time of the mammogram: age, race/ethnicity, age at menarche, age at first birth, body mass index (BMI), history of breast biopsy, history of atypical hyperplasia or benign breast findings, and family history of breast cancer. Breast density measurements were pulled from radiology reports in electronic medical records (EMR) and were classified based on the American College of Radiology’s Breast Imaging-Reporting and Data System (BI-RADS). EMR was also used to supplement missing survey information on the history of breast biopsy, atypical hyperplasia, BMI, and history of prior breast cancer. At MGH, the questionnaire asked whether women had ever had “benign tissue removed from the breast” which was considered as evidence of atypical hyperplasia or benign breast findings. Additionally, at MGH and NWH, we additionally included data from pathology reports on diagnoses of atypical hyperplasia, lobular carcinoma in situ (LCIS), or lobular neoplasia. At UPenn, women were asked whether they had previously been diagnosed with atypical hyperplasia. Additional detail about benign breast conditions was not available for UPenn. Missing BMI specifically was supplemented with the closest measurement from EMR within 1 year prior to or 6 months after the mammogram. The presence of BRCA1 and BRCA2 pathogenic genetic mutations was also collected from linkage with genetic counseling records. We included the first mammogram visit over the time period for each woman, using covariates collected at the time of the first mammogram visit in the cohort for analysis (N = 91,094 at MGH; N = 54,032 at NWH; N = 48,035 at UPenn).
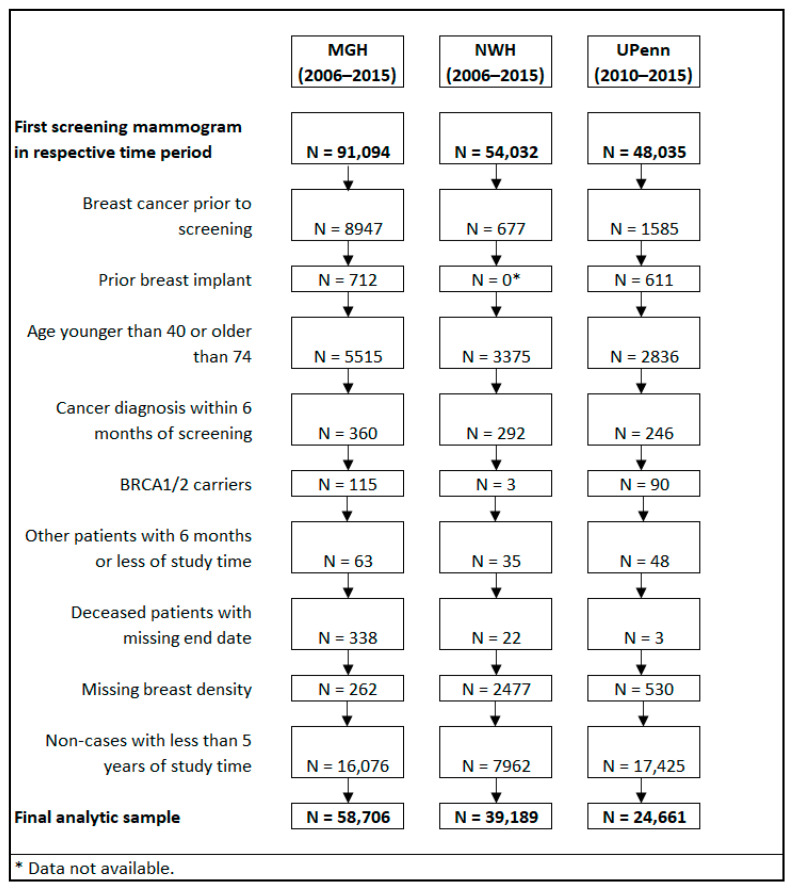
Patients with breast cancer prior to screening were excluded, as were patients diagnosed with cancer within 6 months of screening (Figure 1). For MGH and UPenn, patients with breast implants were excluded; information on breast implants was not available for NWH. Patients who did not fall between the ages of 40 and 84 were excluded, as were those with known pathogenic BRCA1 or BRCA2 mutations since the BCRAT and BCSC models are not appropriate for women with these mutations. Patients with less than 5 years of follow-up time and deceased patients with no date of death or no date of the last contact were also excluded. Patients with missing BI-RADS breast density were excluded. These exclusions resulted in final analytic samples of 58,706 patients from MGH, 39,189 patients from NWH, and 24,661 patients from UPenn (Figure 1).
Figure 1.
Exclusion criteria for screening mammography population by study site.
2.2. Outcomes
Breast cancer cases were determined through to 31 December 2017, using a combination of hospital cancer registries and state health department cancer registries. At MGH and NWH, breast cancer diagnosis information was obtained from the Massachusetts Cancer Registry, while at UPenn, breast cancer diagnosis information was obtained from Pennsylvania, New Jersey, and Delaware state cancer registries. Invasive breast cancers were categorized into molecular subtypes based on the expression of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2). Borderline ER and PR results were considered positive, while borderline HER2 results were considered unknown [12]. Since this analysis predicted a 5-year absolute risk of breast cancer, only patients with invasive breast cancer within 5 years were considered cases. Patients with ductal carcinoma in situ (DCIS) were not considered as cases, since the risk models considered the risk of invasive breast cancer.
2.3. Statistical Analysis
All patients were followed from the date of first screening mammogram visit until breast cancer diagnosis, death, or administrative censoring on 31 December 2017. We estimated the 5-year absolute risk of breast cancer using the BCRAT, BCSC, BRCAPRO, and BRCAPRO+BCRAT models and compared the results with observed breast cancer outcomes. Death was treated as a competing risk. We used the BCRA R package (v2.1) for BCRAT (https://dceg.cancer.gov/tools/risk-assessment/bcra, accessed on 23 November 2021); the BayesMendel R package (v2.1-7) for BRCAPRO and BRCAPRO+BCRAT (https://projects.iq.harvard.edu/bayesmendel/bayesmendel-r-package, accessed on 23 November 2021); and the BCSC SAS program (v2.0) for BCSC (https://tools.bcsc-scc.org/BC5yearRisk/sourcecode.htm, accessed on 23 November 2021).
We assumed patients with missing data on atypical hyperplasia had no atypical hyperplasia (N = 59,285, 48.37%) and that those with a missing number of biopsy examinations had no such examinations (N = 18,549, 15.22%). Regarding missing menopause status, any person over 55 or anyone who had stopped menstruating was categorized as postmenopausal [13]. Anyone who did not meet this criterion was categorized as premenopausal. For patients without, or with incorrect, or ambiguous family history information (approximately 3% total), we assumed that they had no affected relatives.
We validated and compared the performance of BCRAT, BCSC, BRCAPRO, and BRCAPRO+BCRAT with respect to the absolute risk of invasive breast cancer. We also compared these models in subgroups defined by race/ethnicity, invasive molecular subtypes (ER/PR+HER2−, ER/PR+HER2+ or ER/PR−HER2+, or ER/PR/HER2−), a number of affected first- or second-degree relatives, age <50 or ≥50 years, and BMI <30 kg/m2 or ≥30 kg/m2. We performed sensitivity analyses evaluating model performance by site and comparing performance for Black and White women only at UPenn since the majority of Black women were from that site.
Calibration of risk prediction models was evaluated by the observed to the expected ratio (O/E), where the numerator is the observed count of invasive breast cancer cases that arose within 5 years of the initial mammogram, and the denominator is the expected number of cases predicted by the model, obtained by the sum of the predicted 5-year absolute risk estimates. An O/E ratio of 1 indicates perfect calibration, an O/E ratio >1 indicates that the model under-predicts the true number of cases, and an O/E ratio <1 indicates that the model over-predicts the true number of cases. Calibration curves were plotted as the observed proportion of breast cancer cases versus the predicted proportion of breast cancer cases in each decile of predicted absolute risk. Discrimination was assessed by area under the receiver operating characteristic curve (AUC). AUC measures the probability that a prediction model provides higher absolute risk scores for cases than for non-cases. An AUC of 0.5 indicates that the model performs no better than chance, while an AUC of 1 indicates perfect discrimination. The clinically recognized threshold for elevated 5-year risk of breast cancer for chemoprevention is 1.67% [14,15]. This cutoff was used in calculating the true positive rate (TPR) (sensitivity) and the false positive rate (FPR (1— specificity)). TPR was calculated as the proportion of cases who were categorized as high-risk by the model and the FPR was calculated as the proportion of non-cases who were categorized as high-risk by the model.
To obtain 95% confidence intervals (CI) for the performance metrics we generated bootstrap samples and calculated metrics for each model in each bootstrapped sample [16]. To test the statistical significance of observed differences in AUCs across models and strata, we calculated the difference in AUC between two models or two strata for each bootstrap replicate, then calculated the test statistic as the observed difference in AUCs squared, divided by the variance of the difference in AUC derived from the bootstrapped samples. Using this statistic, we obtained p-values for the difference in AUC between pairwise comparisons of models and strata using the Chi-square distribution with 1 degree of freedom. All statistical tests were two-sided using an alpha of 0.05. In addition, we adjusted for multiple comparisons (we performed a total of 34 model and strata comparisons) by the Bonferroni method, with corrected p-values less than 0.00147 considered statistically significant (p = 0.05/34 comparisons).
We also compared the proportion of patients categorized as high vs. low absolute risk, comparing two models at a time and using the cutoff of 1.67% 5 years risk. All analyses were performed using R statistical software (www.R-project.org, accessed on 23 November 2021).
3. Results
Table 1 displays the descriptive characteristics of the study population by site. Most notably, the NWH population was younger than the MGH and UPenn populations, the UPenn population included nearly 46% Black or African American women, and distributions of age at first birth, breast density, and the proportion of missing data differed across sites. Among the 122,556 women in the study population, 1734 were diagnosed with breast cancer within 5 years of their screening mammogram.
Table 1.
Risk factors by study site.
| Risk Factors | MGH | NWH | UPenn |
|---|---|---|---|
| N = 58,706 | N = 39,189 | N = 24,661 | |
| N (%) | |||
| Age, mean +/− SD | 52.57 +/− 9.55 | 51.37 +/− 8.82 | 55.6 +/− 9.29 |
| Age | |||
| 40–44 years | 15,723 (26.78) | 10,796 (27.55) | 3966 (16.08) |
| 45–49 years | 9811 (16.71) | 8380 (21.38) | 3851 (15.62) |
| 50–54 years | 9246 (15.75) | 6976 (17.80) | 4154 (16.84) |
| 55–59 years | 8862 (15.10) | 5245 (13.38) | 4226 (17.14) |
| 60–64 years | 7047 (12.00) | 3845 (9.81) | 3787 (15.36) |
| 65–69 years | 4724 (8.05) | 2400 (6.12) | 2857 (11.59) |
| 70–74 years | 3293 (5.61) | 1547 (3.95) | 1820 (7.38) |
| Race/Ethnicity | |||
| White | 48,072 (81.89) | 35,394 (90.32) | 11,312 (45.87) |
| Black or African American | 3266 (5.56) | 630 (1.61) | 11,279 (45.74) |
| Hispanic | 3177 (5.41) | 392 (1.00) | 560 (2.27) |
| Asian | 3027 (5.16) | 1545 (3.94) | 920 (3.73) |
| Other/Unknown | 1164 (1.98) | 1228 (3.13) | 590 (2.39) |
| Age at menarche | |||
| 7 to 11 years | 10,216 (17.40) | 5895 (15.04) | 4653 (18.87) |
| 12 to 13 years | 29,764 (50.70) | 22,502 (57.42) | 13,190 (53.49) |
| ≤14 years | 15,529 (26.45) | 10,643 (27.16) | 4445 (18.02) |
| Missing | 3197 (5.45) | 149 (0.38) | 2373 (9.62) |
| Age at first birth | |||
| Nulliparous | 15,673 (26.70) | 7297 (18.62) | 5794 (23.49) |
| <20 years | 5721 (9.75) | 1671 (4.26) | 4731 (19.18) |
| 20–24 years | 11,652 (19.85) | 5979 (15.26) | 5001 (20.28) |
| 25–29 years | 11,349 (19.33) | 9923 (25.32) | 4671 (18.94) |
| ≤30 | 12,217 (20.81) | 14,124 (36.04) | 3895 (15.79) |
| Missing | 2094 (3.57) | 195 (0.50) | 569 (2.31) |
| Breast density | |||
| Almost entirely fat | 4342 (7.40) | 755 (1.93) | 2359 (9.57) |
| Scattered fibroglandular tissue | 23,145 (39.43) | 9608 (24.52) | 12,216 (49.54) |
| Heterogeneously dense | 27,112 (46.18) | 22,898 (58.43) | 9162 (37.15) |
| Extremely dense | 4107 (7.0) | 5928 (15.13) | 924 (3.75) |
| Menopausal | |||
| Pre-or peri-menopausal | 24,645 (41.98) | 19,235 (49.08) | 11,971 (48.54) |
| Post-menopausal | 34061 (58.02) | 19,954 (50.92) | 12,690 (51.46) |
| BMI, mean (SD) | 27.21 (6.39) | 26.2 (5.80) | 29.46 (7.40) |
| Prior Breast Biopsy | |||
| None | 57,807 (98.47) | 32,788 (83.67) | 19,764 (80.14) |
| One | 541 (0.92) | 4656 (11.88) | 4259 (17.27) |
| Two or more | 358 (0.61) | 1745 (4.45) | 638 (2.59) |
| Prior atypical hyperplasia/benign breast findings 1 | 849 (1.45) | 204 (0.52) | 91 (0.37) |
| No. of first-degree relatives with breast cancer (%) | |||
| None | 51,037 (86.94) | 32,666 (83.36) | 20,756 (84.17) |
| One | 7093 (12.08) | 5865 (14.97) | 3500 (14.19) |
| Two or more | 576 (0.98) | 658 (1.68) | 405 (1.64) |
| No. of second-degree relatives with breast cancer (%) | |||
| None | 51,285 (87.36) | 28,052 (71.58) | 21,893 (88.78) |
| One | 5818 (9.91) | 7764 (19.81) | 2169 (8.80) |
| Two or more | 1603 (2.73) | 3373 (8.61) | 599 (2.43) |
| No. of first or second degree relatives with ovarian cancer (%) | |||
| None | 58,646 (99.90) | 39,152 (99.91) | 23,644 (95.88) |
| One | 58 (0.10) | 7 (0.02) | 919 (3.73) |
| Two or more | 2 (0.00) | 30 (0.08) | 98 (0.40) |
| 5-year invasive cancer subtype, N (%) |
|||
| ER/PR+HER2− | 619 (57.31) | 379 (56.48) | 318 (55.99) |
| ER/PR+HER2+ | 77 (7.13) | 55 (8.20%) | 30 (5.28) |
| ERPR−HER2+ | 29 (2.69) | 15 (2.24) | 18 (3.17) |
| ERPR−HER2− | 58 (5.37) | 36 (5.37) | 38 (6.69) |
| Invasive cancer, missing subtype | 21 (1.94) | 27 (4.02) | 14 (2.46) |
1 At MGH, patients reported if they’d ever had benign tissue removed from the breast. At both MGH and NWH, we also included pathology report data on atypical hyperplasia, LCIS, or lobular neoplasia. At UPenn, patients self-reported prior atypical hyperplasia.
In the full study population (Table 2), the AUCs for the models ranged from 0.590 for BRCAPRO to 0.617 for the BCSC model. We observed significant differences in the AUCs comparing BCRAT and BCSC (p = 0.010), BCRAT and BRCAPRO (p = 0.007), BCRAT and BRCAPRO+BCRAT (p = 0.040), BCSC and BRCAPRO (p < 0.001), and BRCAPRO and BRCAPRO+BCRAT (p < 0.001). The differences in AUC remained statistically significant after Bonferroni correction for the comparisons of BRCAPRO and BCSC (p < 1 × 10−8) and BRCAPRO and BRCAPRO+BCRAT. O/E ratios ranged from 1.036 for BCRAT to 1.185 for the BCSC model, and the O/E for the BCSC model was further away from 1. The true positive rate ranged from 30.7% for the BCSC model to 37.8% for the BCRAT model, whereas the false positive rate ranged from 18.4% for BCSC to 24.3% for BRCAPRO. The proportion of patients identified as having high 5-year risk differed across models, with 18.6% of patients identified as high risk based on the BCSC model compared with 24.5% based on the BRCAPRO model. Comparable model performance was observed across the three institutions (Appendix A Table A1).
Table 2.
Overall performance of risk models 1.
| Performance Metric | All Sites (N = 1734 Cases & 120,822 Non-Cases) | |||||||
|---|---|---|---|---|---|---|---|---|
| BCRAT | BCSC | BRCAPRO | BRCAPRO+BCRAT | |||||
| Estimate | 95% CI | Estimate | 95% CI | Estimate | 95% CI | Estimate | 95% CI | |
| O/E | 1.036 | 0.989, 1.084 | 1.185 | 1.130, 1.239 | 1.076 | 1.027, 1.125 | 1.063 | 1.014, 1.112 |
| AUC 1 | 0.604 | 0.590, 0.618 | 0.617 | 0.603, 0.630 | 0.590 | 0.578, 0.603 | 0.608 | 0.594, 0.621 |
| TPR | 0.378 | 0.354, 0.402 | 0.307 | 0.284, 0.328 | 0.358 | 0.338, 0.378 | 0.347 | 0.324, 0.368 |
| FPR | 0.238 | 0.235, 0.240 | 0.184 | 0.181, 0.186 | 0.243 | 0.241, 0.245 | 0.217 | 0.215, 0.219 |
| Patients with high 5-year risk, N (%) |
29,396 (23.99) | - | 22,751 (18.56) | - | 30,016 (24.49) | - | 26,830 (21.89) | - |
1 p-values for difference in AUC: BCRAT vs. BCSC p = 0.010, BCRAT vs. BRCAPRO p = 0.007, BCRAT vs. BRCAPRO+BCRAT p = 0.040, BCSC vs. BRCAPRO p < 0.001, BCSC vs. BRCAPRO+BCRAT p = 0.071, BRCAPRO vs. BRCAPRO+BCRAT p < 0.001. Bonferroni corrected p-values remain significant for BCSC vs. BRCAPRO and BRCAPRO vs. BRCAPRO+BCRAT.
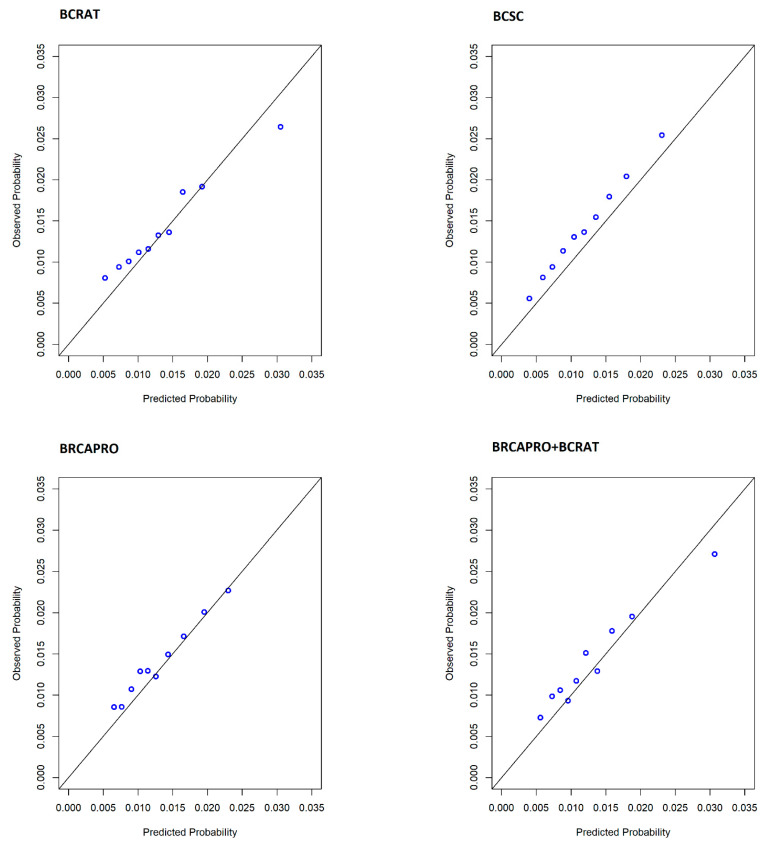
We compared calibration curves for the four models by plotting observed vs. predicted probabilities of breast cancer by deciles (Figure 2). We observed good calibration across deciles for each model. The BCSC model slightly under-predicted the number of cases across all deciles, whereas the BCRAT model under-predicted the number of cases for those in the highest decile.
Figure 2.
Calibration curves for risk prediction models.
Model performance stratified by race/ethnicity is displayed in Table 3. There were 209 breast cancers diagnosed among Black or African American women. Among Black or African American women, the AUCs ranged from 0.610 for BRCAPRO to 0.644 for BCSC, though the differences in AUC by race did not reach statistical significance. In addition, there was no significant difference in AUCs for any model between Black or African American and White women. O/E ratios ranged from 1.054 for BRCAPRO to 1.316 for BCSC. The numbers of cancers among Hispanic (N = 32) and Asian women (N = 54) in the study population were small, so results should be interpreted with caution. In the sensitivity analysis limited to women at UPenn, there was no significant difference in the AUC between Black and White women for the BCRAT or BCRAT+BRCAPRO, but BCSC (p = 0.006) and BRCAPRO (p = 0.049) had significantly higher AUC among Black compared with White women, though the O/E ratio for both Black (O/E = 1.40) and White (O/E = 1.41) women at UPenn indicated under-prediction of cancer cases for BCSC (Appendix C Table A2).
Table 3.
Performance of risk models by race/ethnicity 1.
| Performance Metric by Race/Ethnicity | BCRAT | BCSC | BRCAPRO | BRCAPRO+BCRAT | ||||
|---|---|---|---|---|---|---|---|---|
| White (N = 1411 cases) | Estimate | 95% CI | Estimate | 95% CI | Estimate | 95% CI | Estimate | 95% CI |
| O/E | 1.031 | 0.98, 1.082 | 1.183 | 1.124, 1.242 | 1.100 | 1.045, 1.154 | 1.063 | 1.012, 1.116 |
| AUC | 0.601 | 0.588, 0.613 | 0.607 | 0.595, 0.621 | 0.581 | 0.567, 0.596 | 0.604 | 0.59, 0.616 |
| TPR | 0.410 | 0.387, 0.432 | 0.337 | 0.311, 0.361 | 0.378 | 0.352, 0.404 | 0.377 | 0.351, 0.402 |
| FPR | 0.270 | 0.267, 0.273 | 0.215 | 0.212, 0.217 | 0.269 | 0.266, 0.272 | 0.244 | 0.241, 0.246 |
| Patients with high 5-year risk, N (%) |
25,777 (27.2) | - | 20,503 (21.63) | - | 25,628 (27.04) | - | 23,275 (24.56) | - |
| Black or African American (N = 209 cases) |
||||||||
| O/E | 1.100 | 0.964, 1.24 | 1.316 | 1.155, 1.483 | 1.054 | 0.924, 1.186 | 1.141 | 1.000, 1.287 |
| AUC | 0.614 | 0.581, 0.647 | 0.644 | 0.606, 0.675 | 0.610 | 0.577, 0.646 | 0.617 | 0.585, 0.653 |
| TPR | 0.321 | 0.267, 0.382 | 0.225 | 0.171, 0.282 | 0.378 | 0.319, 0.445 | 0.287 | 0.234, 0.350 |
| FPR | 0.175 | 0.169, 0.181 | 0.103 | 0.098, 0.107 | 0.244 | 0.237, 0.251 | 0.169 | 0.164, 0.175 |
| Patients with high 5-year risk, N (%) |
2689 (17.72) | - | 1584 (10.44) | - | 3738 (24.63) | - | 2588 (17.05) | - |
| Hispanic (N = 32 cases) | ||||||||
| O/E | 0.748 | 0.492, 0.983 | 1.028 | 0.68, 1.354 | 0.840 | 0.552, 1.106 | 0.934 | 0.614, 1.223 |
| AUC | 0.583 | 0.489, 0.699 | 0.582 | 0.491, 0.671 | 0.570 | 0.495, 0.645 | 0.567 | 0.467, 0.668 |
| TPR | 0.063 | 0.000, 0.172 | 0.000 | 0.000, 0.000 | 0.000 | 0.000, 0.000 | 0.000 | 0.000, 0.000 |
| FPR | 0.051 | 0.044, 0.057 | 0.020 | 0.016, 0.024 | 0.006 | 0.004, 0.008 | 0.024 | 0.02, 0.029 |
| Patients with high 5-year risk, N (%) |
209 (5.06) | - | 81 (1.96) | - | 24 (0.58) | - | 100 (2.42) | - |
| Asian (N = 54 cases) | ||||||||
| O/E | 1.588 | 1.172, 1.999 | 1.157 | 0.860, 1.458 | 0.962 | 0.714, 1.212 | 1.054 | 0.781, 1.327 |
| AUC | 0.557 | 0.476, 0.641 | 0.621 | 0.555, 0.705 | 0.617 | 0.542, 0.703 | 0.588 | 0.506, 0.656 |
| TPR | 0.000 | 0.000, 0.000 | 0.037 | 0.000, 0.093 | 0.000 | 0.000, 0.000 | 0.037 | 0.000, 0.093 |
| FPR | 0.021 | 0.018, 0.025 | 0.017 | 0.014, 0.021 | 0.004 | 0.003, 0.006 | 0.068 | 0.062, 0.075 |
| Patients with high 5-year risk, N (%) |
116 (2.11) | - | 96 (1.75) | - | 23 (0.42) | - | 373 (6.79) | - |
1 p-values for comparisons of AUCs for risk models by race/ethnicity are not statistically significant.
Table 4 displays the performance of the risk models by molecular subtype. Given that the models are not trained to predict subtype-specific risk, we did not assess model calibration. AUCs were lower for triple-negative (N = 132, AUC range 0.564–0.585) and HER2+ cancers (N = 224, AUC range 0.513–0.567) compared with ER/PR+HER2− cancers (N = 1316, AUC range 0.605–0.629). The differences in AUCs between ER/PR+HER2− disease and HER2+ disease were statistically significant (all p-values < 0.009), but differences did not reach statistical significance for ER/PR+HER2− compared with ER/PR/HER2− (triple-negative breast cancer, TNBC). After Bonferroni correction, the difference in AUC for BRCAPRO between ER/PR+HER2− and HER2+ disease remained statistically significant (p < 1 × 10−5).
Table 4.
Performance of risk models by molecular subtypes.
| Performance Metric by Subtype | BCRAT 1 | BCSC 2 | BRCAPRO 3 | BRCAPRO+BCRAT 4 | ||||
|---|---|---|---|---|---|---|---|---|
| ER/PR+HER2− (N = 1316 cases) |
Estimate | 95% CI | Estimate | 95% CI | Estimate | 95% CI | Estimate | 95% CI |
| AUC | 0.616 | 0.603, 0.631 | 0.629 | 0.615, 0.645 | 0.605 | 0.590, 0.621 | 0.621 | 0.606, 0.636 |
| TPR | 0.390 | 0.363, 0.417 | 0.313 | 0.290, 0.337 | 0.384 | 0.360, 0.410 | 0.358 | 0.331, 0.384 |
| FPR | 0.238 | 0.236, 0.240 | 0.184 | 0.182, 0.186 | 0.243 | 0.241, 0.245 | 0.217 | 0.215, 0.219 |
| Patients with high 5-year risk, N (%) | 29,253 (23.95) | - | 22,630 (18.53) | - | 29,902 (24.48) | - | 26,699 (21.86) | - |
| All HER2+ (N = 224 cases) |
||||||||
| AUC | 0.560 | 0.525, 0.600 | 0.567 | 0.535, 0.610 | 0.513 | 0.479, 0.553 | 0.561 | 0.526, 0.599 |
| TPR | 0.330 | 0.270, 0.390 | 0.281 | 0.236, 0.338 | 0.246 | 0.200, 0.307 | 0.299 | 0.248, 0.353 |
| FPR | 0.238 | 0.235, 0.240 | 0.184 | 0.182, 0.186 | 0.243 | 0.241, 0.246 | 0.217 | 0.215, 0.219 |
| Patients with high 5-year risk, N (%) | 28,794 (23.80) | - | 22,266 (18.40) | - | 29,438 (24.33) | - | 26,278 (21.72) | - |
| ER/PR/HER2− (N = 132 cases) |
||||||||
| AUC | 0.570 | 0.516, 0.617 | 0.585 | 0.546, 0.630 | 0.564 | 0.522, 0.604 | 0.569 | 0.513, 0.621 |
| TPR | 0.348 | 0.252, 0.430 | 0.295 | 0.215, 0.375 | 0.295 | 0.216, 0.369 | 0.303 | 0.220, 0.380 |
| FPR | 0.238 | 0.236, 0.240 | 0.184 | 0.182, 0.186 | 0.243 | 0.241, 0.246 | 0.217 | 0.215, 0.219 |
| Patients with high 5-year risk, N (%) | 28,786 (23.80) | - | 22,257 (18.40) | - | 29,435 (24.34) | - | 26,268 (21.72) | - |
1p-values for differences in AUCs for BCRAT: ER/PR+HER2− vs. HER2+ p = 0.008, ER/PR+HER2− vs. TNBC p = 0.084. 2 p-values for differences in AUCS for BCSC: ER/PR+HER2− vs. HER2+ p = 0.002, ER/PR+HER2− vs. TNBC p = 0.081. 3 p-values for differences in AUCS for BRCAPRO: ER/PR+HER2− vs. HER2+ p < 0.001, ER/PR+HER2− vs. TNBC p = 0.100. 4 p-values for differences in AUCS for BRCAPRO+BCRAT: ER/PR+HER2− vs. HER2+ p = 0.003, ER/PR+HER2− vs. TNBC p = 0.063.
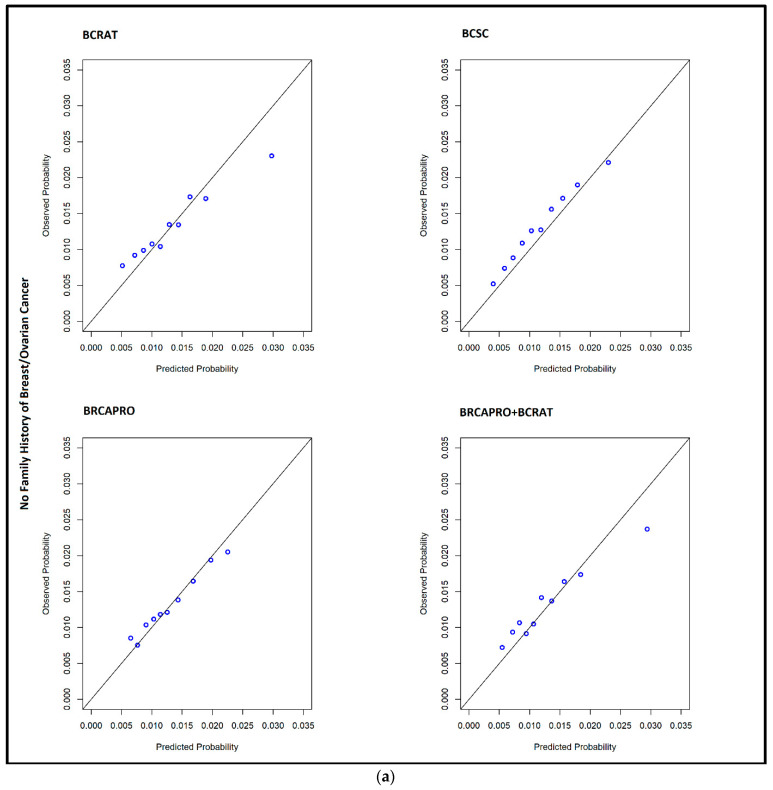
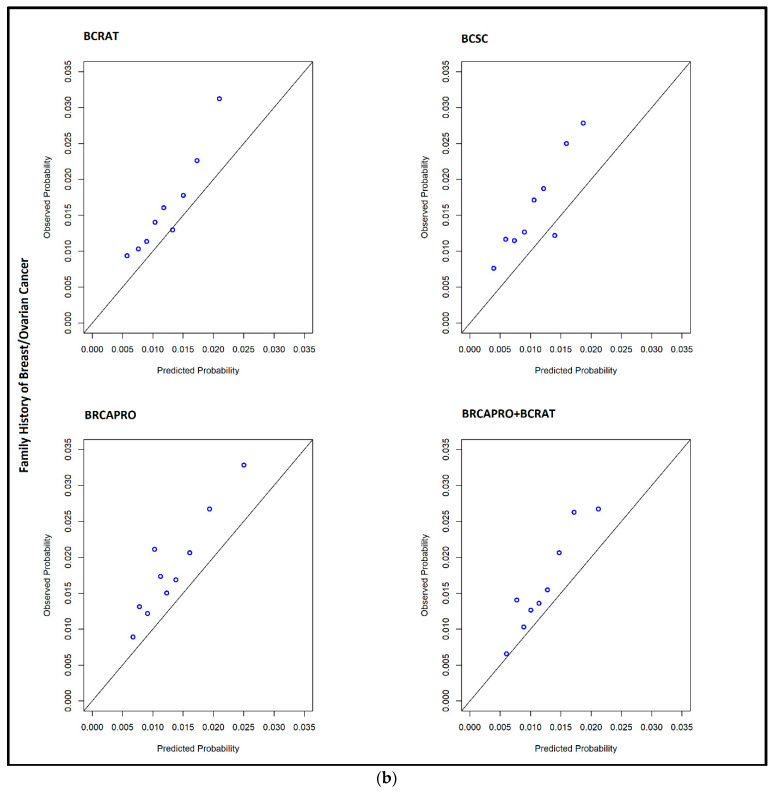
Table 5 displays model performance stratified by family history of breast cancer, age, and BMI. For women with a family history, breast cancers were under-predicted for all models (O/E range 1.263–1.509). AUC estimates were significantly higher for patients with family history than without family history for the BCRAT (p = 0.019) and BRCAPRO+BCRAT (p = 0.016) models. These differences were no longer statistically significant after the Bonferroni correction. O/E ratios for all models among patients with a family history were further from 1 than those without a family history, and confidence intervals did not overlap, suggesting cases were underpredicted for patients with a family history (Appendix B Figure A1). For women under 50, O/E ratios indicated under-prediction of cancers. AUCs did not differ significantly by age. For BMI, all four models had significantly higher AUCs among obese compared with non-obese women (BCRAT p = 0.004, BCSC p < 0.001, BRCAPRO p = 0.001, BRCAPRO+BCRAT p = 0.012). Both the BCSC model and the BRCAPRO model had significantly higher AUC among obese women compared with non-obese women after adjusting for multiple comparisons (BCSC p = 0.000025, BRCAPRO p = 0.0012). However, the under-prediction was more severe for BCSC and BRCAPRO among women with high BMI compared with lower BMI.
Table 5.
Performance of risk models by family history of breast cancer, age, and BMI.
| Family History 1 | No Family History of Breast Cancer (N = 1340 Cases) | |||||||
|---|---|---|---|---|---|---|---|---|
| BCRAT | BCSC | BRCAPRO | BRCAPRO+BCRAT | |||||
| Estimate | 95% CI | Estimate | 95% CI | Estimate | 95% CI | Estimate | 95% CI | |
| O/E | 0.982 | 0.934, 1.045 | 1.115 | 1.061, 1.186 | 1.007 | 0.959, 1.07 | 1.016 | 0.966, 1.081 |
| AUC | 0.594 | 0.582, 0.61 | 0.612 | 0.601, 0.625 | 0.588 | 0.575, 0.601 | 0.597 | 0.584, 0.613 |
| TPR | 0.352 | 0.328, 0.377 | 0.284 | 0.262, 0.311 | 0.361 | 0.339, 0.385 | 0.316 | 0.293, 0.342 |
| FPR | 0.231 | 0.229, 0.235 | 0.179 | 0.177, 0.181 | 0.248 | 0.245, 0.251 | 0.208 | 0.205, 0.211 |
| High 5-year risk, N (%) |
23,590 (23.3) | - | 18,253 (18.03) | - | 25,270 (24.96) | - | 21,173 (20.92) | - |
| Family History of Breast Cancer (N = 394 cases) | ||||||||
| O/E | 1.273 | 1.161, 1.42 | 1.509 | 1.38, 1.686 | 1.402 | 1.283, 1.569 | 1.263 | 1.153, 1.408 |
| AUC | 0.633 | 0.607, 0.659 | 0.631 | 0.603, 0.657 | 0.597 | 0.571, 0.625 | 0.636 | 0.611, 0.666 |
| TPR | 0.467 | 0.42, 0.519 | 0.386 | 0.337, 0.428 | 0.345 | 0.295, 0.392 | 0.454 | 0.405, 0.501 |
| FPR | 0.269 | 0.262, 0.275 | 0.208 | 0.202, 0.213 | 0.220 | 0.213, 0.227 | 0.262 | 0.256, 0.268 |
| High 5-year risk, N (%) |
5806 (27.22) | - | 4498 (21.09) | - | 4746 (22.25) | - | 5657 (26.53) | - |
| Age 2 | <50 (N = 567 cases) | |||||||
| O/E | 1.150 | 1.067, 1.251 | 1.386 | 1.287, 1.508 | 1.219 | 1.134, 1.325 | 1.144 | 1.062, 1.244 |
| AUC | 0.590 | 0.568, 0.616 | 0.617 | 0.593, 0.642 | 0.566 | 0.544, 0.594 | 0.592 | 0.569, 0.62 |
| TPR | 0.136 | 0.11, 0.161 | 0.067 | 0.048, 0.088 | 0.019 | 0.009, 0.03 | 0.129 | 0.102, 0.153 |
| FPR | 0.055 | 0.053, 0.057 | 0.023 | 0.021, 0.024 | 0.009 | 0.008, 0.01 | 0.052 | 0.051, 0.054 |
| High 5-year risk, N (%) |
2947 (5.61) | - | 1216 (2.31) | - | 463 (0.88) | - | 2788 (5.31) | - |
| ≥50 (N = 1167 cases) | ||||||||
| O/E | 0.989 | 0.929, 1.034 | 1.107 | 1.041, 1.16 | 1.018 | 0.958, 1.064 | 1.028 | 0.966, 1.075 |
| AUC | 0.582 | 0.564, 0.598 | 0.595 | 0.579, 0.608 | 0.571 | 0.555, 0.586 | 0.588 | 0.57, 0.603 |
| TPR | 0.496 | 0.467, 0.524 | 0.424 | 0.396, 0.449 | 0.522 | 0.496, 0.549 | 0.453 | 0.424, 0.479 |
| FPR | 0.376 | 0.372, 0.379 | 0.306 | 0.302, 0.309 | 0.420 | 0.417, 0.424 | 0.341 | 0.338, 0.345 |
| High 5-year risk, N (%) |
26,449 (37.77) | - | 21,535 (30.75) | - | 29,553 (42.20) | - | 24,042 (34.33) | - |
| Body Mass Index 3 | BMI < 30 kg/m2 (N = 999 cases) | |||||||
| O/E | 1.080 | 1.019, 1.15 | 1.188 | 1.122, 1.265 | 1.128 | 1.066, 1.2 | 1.102 | 1.040, 1.173 |
| AUC | 0.588 | 0.569, 0.604 | 0.597 | 0.58, 0.612 | 0.568 | 0.552, 0.586 | 0.594 | 0.576, 0.613 |
| TPR | 0.353 | 0.328, 0.382 | 0.305 | 0.277, 0.334 | 0.309 | 0.289, 0.339 | 0.327 | 0.300, 0.358 |
| FPR | 0.236 | 0.233, 0.239 | 0.205 | 0.202, 0.209 | 0.233 | 0.23, 0.237 | 0.217 | 0.213, 0.220 |
| High 5-year risk, N (%) |
16,254 (23.76) | - | 14,157 (20.7) | - | 16,048 (23.46) | - | 14,934 (21.83) | - |
| BMI ≥ 30 kg/m2 (N = 428 cases) | ||||||||
| O/E | 1.164 | 1.054, 1.264 | 1.525 | 1.384, 1.661 | 1.179 | 1.071, 1.285 | 1.206 | 1.091, 1.309 |
| AUC | 0.634 | 0.612, 0.657 | 0.661 | 0.639, 0.683 | 0.617 | 0.592, 0.639 | 0.634 | 0.609, 0.659 |
| TPR | 0.428 | 0.388, 0.468 | 0.250 | 0.215, 0.283 | 0.432 | 0.389, 0.477 | 0.390 | 0.348, 0.435 |
| FPR | 0.231 | 0.226, 0.236 | 0.111 | 0.107, 0.114 | 0.267 | 0.26, 0.273 | 0.212 | 0.208, 0.218 |
| High 5-year risk, N (%) |
6268 (23.42) | - | 3032 (11.33) | - | 7212 (26.94) | - | 5755 (21.5) | - |
1p-value for difference in AUCs for women with and without family history BCRAT p = 0.019, BRCAPRO+BCRAT p = 0.016. No significant associations after Bonferroni correction. 2 No statistically significant differences in AUCs for risk models by age. 3 p-value for difference in AUCs between women with BMI < 30 kg/m2 and women with BMI ≥ 30 kg/m2: BCRAT p = 0.004, BCSC p < 0.001, BRCAPRO p = 0.001, BRCAPRO+BCRAT = 0.012. Differences for BCSC and BRCAPRO remained statistically significant after Bonferroni correction.
Table 6 displays a cross-tabulation of high (≥1.67% 5 years risk) versus low absolute risk classification for all possible pairs of models. Overall, the proportion of discordant classifications ranged from 2.9% for the comparison of BCRAT with BRCAPRO+BCRAT (0.5% + 2.4%) to 19.7% for the comparison of BCSC with BRCAPRO (13.6% + 6.1%). BCSC classified more patients as low risk than other models. Nearly 10% of patients were classified as high risk with the BCRAT model but low risk with BCSC, whereas only 5.7% of patients were classified as low risk on BCRAT and high risk on BCSC. Almost 14% of patients classified as high risk by BRCAPRO were classified as low risk by BCSC, while only 6.1% of patients considered a low risk on BRCAPRO were considered high risk with BCSC. Comparing BCRAT and BRCAPRO, 7.8% of women who were high risk on BCRAT were classified as low risk by BRCAPRO, while 11.1% of women who were high risk on BRCAPRO were classified as low risk by BCRAT. The proportions of patients classified differently by the BRCAPRO+BCRAT model compared with the BCRAT model were smaller.
Table 6.
Concordance between 5-year high-risk estimates across models.
| Risk Model | Risk Level | BCSC | BRCAPRO | BRCAPRO+BCRAT | |||
|---|---|---|---|---|---|---|---|
| N (% of Total) | N (% of Total) | N (% of Total) | |||||
| High | Low | High | Low | High | Low | ||
| BCRAT | High | 15,995 (13.05) | 13,401 (10.93) | 17,880 (14.59) | 11,516 (9.4) | 25,826 (21.07) | 3570 (2.91) |
| Low | 6756 (5.51) | 86,404 (70.5) | 12,136 (9.9) | 81,024 (66.11) | 1004 (0.82) | 92,156 (75.2) | |
| BCSC | High | - | - | 14,297 (11.67) | 8454 (6.9) | 15,013 (12.25) | 7738 (6.31) |
| Low | - | - | 15,719 (12.83) | 84,086 (68.61) | 11,817 (9.64) | 87,988 (71.79) | |
| BRCAPRO | High | - | - | - | - | 17,537 (14.31) | 12,479 (10.18) |
| Low | - | - | - | - | 9293 (7.58) | 83,247 (67.93) | |
4. Discussion
Our study is one of the largest to examine the validity of multiple breast cancer risk prediction models simultaneously in a large and diverse sample of women undergoing mammography screening, allowing comparisons of model performance by race/ethnicity, tumor molecular subtypes, family history, and obesity. We found that model performance was comparable across the four models examined, with moderate discriminatory accuracy, and generally good calibration, consistent with previous estimates [7,17,18,19,20,21,22,23,24,25,26,27,28,29,30]. The BCSC model had the highest AUC but under-predicted the number of cancer cases to a greater degree than the other models. We found no evidence of poorer model performance for Black or African American women compared with White women. AUC estimates were consistently higher for Black or African American women compared to White women, though not statistically significantly different, except among women at UPenn, where the AUC was better for BCSC and BRCAPRO models for Black compared with White women. With the exception of the BRCAPRO model, the O/E ratio was further from 1 for Black or African American women compared to White women, indicating that models under-predicted the number of cases among Black or African American women. Model discriminatory accuracy was poorer for HER2+ than ER/PR+HER2− disease across all models, and also lower for TNBC than ER/PR+HER2− disease, however, the difference was not statistically significant for TNBC. This is the first study to validate the combined BRCAPRO+BCRAT model, which has significantly better discriminatory accuracy than BRCAPRO in this screening population. BCRAT and BRCAPRO+BCRAT had better AUC among women with family history than women without a family history, though O/E ratios were further from 1 for women with family history compared to those without. Across all models, AUCs were higher for obese women than non-obese women, but the calibration was poorer, with under-prediction of cases among obese women. While overall there was good concordance in the patients identified as high and low absolute risk, we found anywhere from 2.4% to 19.7% discordance in pairwise comparisons of the proportions of patients identified as high versus low risk across the models. These findings add to the literature and provide useful performance metrics to consider when selecting breast cancer risk assessment models, and also highlight the complications when applying risk models clinically and the need for improved risk assessment tools.
The measures of calibration and discrimination of the models observed in our study are generally consistent with existing literature [7,17,18,19,20,21,22,23,24,25,26,27,28,29,30]. This suggests that there is no clear winner in terms of which risk model to use for the purpose of directing screening. However, while we found that the models largely agreed on which patients were high risk, there was variation across models. Interestingly, the BCSC model was more likely to classify women as having low risk when compared with the BCRAT model and the BRCAPRO model. While the choice of model may have only marginal differences at the population level, our results highlight that the choice of model may lead to a different assessment of high-risk status for individual women. In the case of supplemental screening, different recommendations would be made if only one model is used compared to if multiple models are used, and an individual is considered high-risk based on any of the applied models. If the goal is to increase the number of women identified as high risk, we might take the latter approach. If the goal is to minimize the use of resources and unnecessary procedures, we may use only one model or set a higher threshold for high-risk status. Answering such questions is key to implementing a risk-based approach to screening.
Our study found no significant difference in model performance between Black or African American and White women. To our knowledge, ours is the first study to compare the performance of multiple breast cancer risk prediction models simultaneously for White and Black women. AUCs tended to be higher for Black or African American than White women, and calibration tended to be poorer for Black or African American than White women, though p-values for differences in AUCs were not significant, and confidence intervals for O/E ratios for Black or African American and White women were overlapping. Despite our large population, there were only 209 breast cancers diagnosed among Black or African American women, which may limit our power to detect differences. The original BCRAT model was found to underestimate risk among Black or African American women and was therefore updated to better predict breast cancer risk for Black or African American women based on the results of the Women’s Contraceptive and Reproductive Experiences (CARE) study [5]. However, even after this update, the BCRAT model was still shown to significantly underestimate risk in a study that included 725 cases and 725 controls from the Black Women’s Health Study [31]. Validation of the BCSC model among women undergoing mammography screening showed that risk was under-predicted for non-Hispanic Black women, though the confidence interval of the observed to expected ratio overlapped with that for White women [28]. The AUC of the BCSC model among Black women was not reported in this study. While discrimination and calibration were comparable across models for Black or African American and White women in our study, all risk models identified a smaller proportion of Black or African American than White women as high risk. For example, over 21% of White women were identified as having high 5 years risk based on the BCRAT model, compared with 10% of Black or African American women. This illustrates the fact that using existing risk models to direct supplemental screening will result in fewer Black or African American women than White women qualifying for additional imaging. While historically Black or African American women in the U.S. have had lower breast cancer mortality than White women, breast cancer incidence has increased for Black or African American women over the past 20 years, with incidence rates now very close to those of White women (127.3 vs. 131.6 per 100,000 women) [1]. Future research should closely evaluate the potential effects of using risk models to guide prevention measures on racial disparities in breast cancer.
Our results further suggest that risk models do not identify the risk of triple-negative or HER2+ tumors as well as hormone receptor-positive HER2− tumors. Our results are consistent with an analysis of the Women’s Health Initiative, which showed that the BCRAT model predicted ER+ breast cancers, but not ER− breast cancers [21] and with our prior analysis with a smaller sample [11]. Given that HER2+ and TNBCs are more aggressive, future studies should attempt to build subtype-specific risk prediction models. Identification of risk of poor prognosis cancers would allow targeting of prevention strategies such as intensive screening to women at greatest risk for breast cancer death. Thus far, the lower prevalence of both HER2+ and triple-negative tumors have limited the ability to generate subtype-specific models.
BRCAPRO and BRCAPRO+BCRAT use more extensive family history data than the BCSC and BCRAT models. In this screening cohort, approximately 20% of patients had any family history of breast cancer, including first or second-degree relatives. We found that BRCAPRO+BCRAT had significantly better AUC than BRCAPRO and calibration was very similar for both models overall, and among the subset of women with family history. This highlights the need to balance between a potentially small increased accuracy of risk assessment including extended family history, and the additional time and computational burden of collecting and analyzing extensive family history data. It is important to note that detailed family history assessment has the added benefit of identifying patients who may be at high risk for high and moderate penetrance mutations such as BRCA1/2 who may benefit from genetic testing.
We found significantly higher AUC across models among obese women compared with non-obese women, though calibration was poorer for obese women, particularly for the BCSC model. While none of the existing models include BMI as a predictor, the BCSC model does incorporate breast density, which is highly correlated with BMI, with women with higher BMI having lower breast density on average. Therefore, the BCSC model likely downgrades risk for many obese women given their lower breast density, leading to under-prediction of risk. Breast density as coded by the radiologist has been shown to have poor reproducibility [32]. Novel measures of breast density, such as quantitative measures of volumetric breast density from digital breast tomosynthesis may prove a better risk marker than radiologist-coded breast density, particularly for obese women. A recent study found that the effect of volumetric breast density on breast cancer risk was strongest in overweight and obese women [33], suggesting that risk models may need to incorporate these novel measures and include interaction terms between density and BMI in order to improve risk assessment for obese women. Given that the prevalence of obesity is large and increasing, and that obese women are more likely to be diagnosed with more aggressive diseases [34,35], the inclusion of BMI in risk models should be explored, with a closer evaluation of how to best predict risk among women with high BMI.
To our knowledge, this is one of the largest studies to compare multiple breast cancer risk models among women undergoing mammography screening in the U.S. The large and diverse cohort enabled comparisons of model performance by race/ethnicity, molecular subtypes, family history, age, and BMI. Additionally, we were able to evaluate a novel combination of the BRCAPRO and BCRAT model, BRCAPRO+BCRAT, that may be useful for patients with an extensive family history of breast and ovarian cancers. Our findings provide directions for future research by identifying subsets of the population for which existing models may have suboptimal performance.
Several limitations should be noted when interpreting the findings. We were unable to include the IBIS/Tyrer-Cuzick model because we lacked sufficient data on the use of menopausal hormone therapy for input to this model. Due to the small number of Hispanic and Asian/Pacific Islander patients, there were too few cancer outcomes to make meaningful inferences on the performance of the risk models in these racial/ethnic groups. There were 209 breast cancers among Black or African American women, which is also a relatively small sample. Most of the Black or African American patients were screened at one of the three sites, and therefore results for Black or African American patients may be confounded by site-specific measurement biases. In addition, the data on prior biopsy from MGH was incomplete. However, we believe the data on atypical hyperplasia to be reasonably accurate since it incorporated both self-reported data and information extracted from biopsy reports. However, we lacked data on other benign breast conditions, such as non-proliferative benign breast diseases and proliferative benign breast diseases without atypia. While the BCRAT uses atypical hyperplasia, the BCSC model uses benign breast disease, and therefore we may be underestimating risk among patients since we do not have full data on benign breast diseases. This highlights the broader concern, that there is variation in data quality across risk factors and across sites. However, this is the reality when data collected for clinical purposes is utilized for risk assessment. The inclusion of three cohorts with slightly different risk collection instruments may help in reducing the effect of misclassification of risk factors at any one site on the results. We treated patients diagnosed with DCIS as non-cases since the risk models considered focus on the risk of invasive breast cancer. However, since risk factors for DCIS and invasive cancer share common risk factors, this may have led to the poorer observed performance of our models. Additionally, the purpose of this study was to evaluate risk models that would be feasible to integrate into clinical care in mammography to support decision-making in the general population. We excluded known BRCA1/2 carriers because the BCRAT and BCSC risk models are specifically designed to predict risk in non-carriers. There were fewer BRCA1/2 carriers at NWH than MGH and UPenn, which is partly expected, as NWH is a community hospital, whereas MGH and UPenn have large cancer genetics clinics. It is also possible that BRCA1/2 mutation status was less well annotated in health records at NWH, and we may have included some BRCA1/2 mutation carriers in the analysis, which may bias results in unknown ways. However, given the small prevalence of BRCA1/2 mutations in the general population, we expect the magnitude of this bias to be small. Finally, we lacked genetic data to incorporate polygenic risk scores into risk models, which has been shown to improve predictive accuracy. A recent study showed that adding the 313 SNP polygenic risk score to classical breast cancer risk factors improved the AUC from 56% to 64% among women younger than 50 and from 57% to 64% among women aged 50 and older [36]. Polygenic risk scores are not yet integrated into clinical practice to direct supplemental screening, and further studies are needed to direct the implementation of such approaches. Finally, we performed a large number of comparisons, and therefore some of the statistically significant associations may have been the result of chance rather than true underlying differences in performance. We provide Bonferroni corrected p-values and found several associations that remained statistically significant after Bonferroni correction.
5. Conclusions
In summary, our research provides important data on the performance of breast cancer risk assessment models among women undergoing mammography screening in the U.S. and provides important data on how models perform in different subsets of the population, including Black or African American women and women with a family history of breast cancer, and how well the models predict breast cancer subtypes. We validate the BRCAPRO+BCRAT model which may be beneficial for use among women with a family history of breast and ovarian cancer. These findings are useful to develop risk assessment strategies but ultimately point to the need to further improve risk prediction by incorporating additional risk factors, such as quantitative measures of breast density and genetic markers.
Appendix A
Table A1.
Performance of risk models by site.
| Performance Metric by Site | BCRAT | BCSC | BRCAPRO | BRCAPRO+BCRAT | ||||
|---|---|---|---|---|---|---|---|---|
| MGH (N = 804 cases) | Estimate | 95% CI | Estimate | 95% CI | Estimate | 95% CI | Estimate | 95% CI |
| O/E | 1.060 | 0.993, 1.131 | 1.196 | 1.119, 1.275 | 1.057 | 0.988, 1.126 | 1.095 | 1.025, 1.166 |
| AUC | 0.595 a | 0.576, 0.615 | 0.620 c | 0.604, 0.639 | 0.587 d | 0.567, 0.610 | 0.603 b | 0.585, 0.625 |
| TPR | 0.331 | 0.302, 0.362 | 0.270 | 0.244, 0.307 | 0.340 | 0.311, 0.375 | 0.294 | 0.263, 0.325 |
| FPR | 0.204 | 0.201, 0.207 | 0.162 | 0.159, 0.166 | 0.238 | 0.235, 0.241 | 0.186 | 0.183, 0.188 |
| Patients with high 5-year risk, N (%) |
12,089 (20.59) | - | 9600 (16.35) | - | 14,049 (23.93) | - | 10,983 (18.71) | - |
| NWH (N = 512 cases) | ||||||||
| O/E | 0.930 | 0.850, 1.003 | 1.039 | 0.950, 1.123 | 1.026 | 0.938, 1.107 | 0.947 | 0.866, 1.022 |
| AUC | 0.618 | 0.592, 0.640 | 0.624 c | 0.602, 0.646 | 0.599 d | 0.574, 0.622 | 0.622 | 0.598, 0.644 |
| TPR | 0.408 | 0.368, 0.452 | 0.369 | 0.329, 0.406 | 0.324 | 0.281, 0.365 | 0.387 | 0.345, 0.433 |
| FPR | 0.252 | 0.248, 0.256 | 0.218 | 0.213, 0.222 | 0.205 | 0.201, 0.209 | 0.228 | 0.224, 0.232 |
| Patients with high 5-year risk, N (%) |
9960 (25.42) | - | 8603 (21.95) | - | 8078 (20.61) | - | 9015 (23.00) | - |
| UPenn (N = 418 cases) | ||||||||
| O/E | 1.146 | 1.044, 1.261 | 1.402 | 1.275, 1.544 | 1.186 | 1.080, 1.306 | 1.174 | 1.070, 1.294 |
| AUC | 0.598 e | 0.567, 0.625 | 0.603 c | 0.578, 0.631 | 0.576 d | 0.549, 0.603 | 0.597 | 0.568, 0.624 |
| TPR | 0.433 | 0.380, 0.477 | 0.304 | 0.265, 0.344 | 0.433 | 0.389, 0.475 | 0.402 | 0.356, 0.447 |
| FPR | 0.296 | 0.291, 0.301 | 0.182 | 0.177, 0.187 | 0.318 | 0.313, 0.323 | 0.275 | 0.270, 0.280 |
| Patients with high 5-year risk, N (%) |
7347 (29.79) | - | 4548 (18.44) | - | 7889 (31.99) | - | 6832 (27.70) | - |
a p < 0.05 for Gail AUC vs. BCSC within same site. b p < 0.05 for Gail AUC vs. BRCAGAIL AUC within same site. c p < 0.05 for BCSC AUC vs. BRCAPRO AUC within same site. d p < 0.05 for BRCAPRO AUC vs. BRCAGAIL AUC within same site. e p < 0.05 for Gail AUC vs. BRCAPRO AUC within same site.
Appendix B
Figure A1.
(a) Calibration curves for four models for individuals with no family history of breast cancer. (b) Calibration curves for four models for individuals with a family history of breast cancer. Each point on the figure represents the observed probability of breast cancer (Y-axis) for each decile of probability of breast cancer predicted by the model (X-axis). The diagonal line indicates perfect agreement between predicted and observed probability of breast cancer.
Appendix C
Table A2.
Performance of risk models by race/ethnicity at UPenn.
| Performance Metric by Race | BCRAT | BCSC | BRCAPRO | BRCAPRO+BCRAT | ||||
|---|---|---|---|---|---|---|---|---|
| White (N = 220 Cases) | Estimate | 95% CI | Estimate | 95% CI | Estimate | 95% CI | Estimate | 95% CI |
| O/E | 1.129 | 0.999, 1.281 | 1.400 | 1.241, 1.588 | 1.259 | 1.117, 1.434 | 1.161 | 1.027, 1.317 |
| AUC | 0.580 | 0.544, 0.619 | 0.565 a | 0.525, 0.601 | 0.549 b,c | 0.513, 0.583 | 0.583 | 0.548, 0.623 |
| TPR | 0.532 | 0.47, 0.595 | 0.373 | 0.309, 0.431 | 0.473 | 0.402, 0.533 | 0.500 | 0.436, 0.561 |
| FPR | 0.421 | 0.412, 0.429 | 0.273 | 0.264, 0.281 | 0.401 | 0.394, 0.41 | 0.383 | 0.373, 0.391 |
| Patients with high 5-year risk, N (%) |
4783 (42.28) | - | 3112 (27.51) | - | 4557 (40.28) | - | 4354 (38.49) | - |
| Black or African American (N = 170 cases) d |
||||||||
| O/E | 1.152 | 0.984, 1.377 | 1.413 | 1.218, 1.691 | 1.112 | 0.957, 1.33 | 1.190 | 1.016, 1.423 |
| AUC | 0.610 | 0.571, 0.653 | 0.644 | 0.594, 0.684 | 0.608 | 0.565, 0.648 | 0.615 | 0.572, 0.659 |
| TPR | 0.353 | 0.276, 0.424 | 0.241 | 0.178, 0.305 | 0.429 | 0.353, 0.5 | 0.324 | 0.263, 0.393 |
| FPR | 0.199 | 0.192, 0.206 | 0.110 | 0.105, 0.116 | 0.273 | 0.265, 0.282 | 0.193 | 0.186, 0.2 |
| Patients with high 5-year risk, N (%) |
2266 (20.09) | - | 1267 (11.23) | - | 3108 (27.56) | - | 2201 (19.51) | - |
a p < 0.05 between White vs. Black women for BCSC model. b p < 0.05 between White vs. Black women for BRCAPRO model. c p < 0.05 between BRCAPRO vs. BRCAPRO+BRAT models for White women. d p < 0.05 between BRCAT vs. BCSC, BCSC vs. BCRAPRO, & BCSC vs. BRCAPRO+BRCAT for Black women.
Author Contributions
Conceptualization, A.M.M.; methodology, A.M.M., J.C., G.P., D.B.; software, G.P., D.B., Z.G.; formal analysis, Y.L., J.L., T.H., Z.G.; resources, A.M.M., K.A., G.P., J.C., K.H., C.L., A.S.; data curation, A.M.M., Y.L., S.E.; writing—original draft preparation, A.M.M., Y.L., S.E.; writing—review and editing, A.M.M., Y.L., S.E., Z.G., J.L., T.H., K.H., A.S., D.K., E.C., C.L., K.A., D.B., G.P., J.C.; visualization, Y.L.; supervision, A.M.M.; project administration, A.M.M.; funding acquisition, A.M.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the American Cancer Society (PI McCarthy 131052-MRSG-17-144-01-CCE) and the National Cancer Institute (PI Chen R01-CA236468).
Institutional Review Board Statement
This study was deemed exempt from review by the University of Pennsylvania Institutional Review Board.
Informed Consent Statement
Patient consent was waived due to this study being a retrospective analysis of existing data, and therefore posing no more than minimal risk to patients.
Data Availability Statement
The data underlying this article cannot be shared publicly in order to protect patient privacy. The data may be shared in a de-identified format on reasonable request to the corresponding author.
Conflicts of Interest
Giovanni Parmigiani is a co-founder and equity holder in Phaeno Biotechnologies, a member of the Scientific Advisory Board of Konica-Minolta Precision Medicine (which includes Ambry Genetics and Invicro), and a consultant for Delfi Diagnostics and Foundation Medicine. Danielle Braun and Giovanni Parmigiani co-lead the BayesMendel lab, which develops and maintains the BayesMendel software package. This includes a variety of risk assessment tools including BRCAPRO, PancPRO, MelaPRO, MMRpro, and PanelPRO and is licensed for commercial use. All licensing revenues are used for software maintenance and upgrades. Neither BayesMendel lab leaders nor members derive personal income from BayesMendel licenses. Danielle Braun and Giovanni Parmigiani are co-inventor of the Ask2me tool, which is commercially licensed. Kevin Hughes receives honoraria from Hologic (surgical implant for radiation planning with breast conservation and wire free breast biopsy) and Myriad Genetics, Hughes has financial interests in CRA Health (Formerly Hughes RiskApps) which recently was sold to Volpara. CRA Health develops risk assessment models/software with a particular focus on breast cancer and colorectal cancer. Hughes is a founder of the company. Hughes is the Co-Creator of Ask2Me.Org which is freely available for clinical use and is licensed for commercial use by the Dana Farber Cancer Institute and the MGH. Hughes’s interests in CRA Health and Ask2Me.Org were reviewed and are managed by Massachusetts General Hospital and Partners Health Care in accordance with their conflict-of-interest policies. Constance Lehman is co-founder of Clairity, which is developing an AI-based risk assessment products. Lehman’s interests in Clairity were reviewed and are managed by Massachusetts General Hospital and Partners Health Care in accordance with their conflict-of-interest policies. Emily Conant is on the grant and advisory boards for iCAD, Inc. and Hologic, Inc. The remaining authors have no conflicts to disclose. The funders of this study had no role in its design; the collection, analyses, or interpretation of the data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Cintolo-Gonzalez J.A., Braun D., Blackford A.L., Mazzola E., Acar A., Plichta J.K., Griffin M., Hughes K.S. Breast cancer risk models: A comprehensive overview of existing models, validation, and clinical applications. Breast Cancer Res. Treat. 2017;164:263–284. doi: 10.1007/s10549-017-4247-z. [DOI] [PubMed] [Google Scholar]
- 3.Banegas M.P., John E.M., Slattery M.L., Gomez S.L., Yu M., LaCroix A.Z., Pee D., Chlebowski R.T., Hines L.M., Thompson C.A., et al. Projecting Individualized Absolute Invasive Breast Cancer Risk in US Hispanic Women. J. Natl. Cancer Inst. 2017;109:djw215. doi: 10.1093/jnci/djw215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gail M.H., Brinton L.A., Byar D.P., Corle D.K., Green S.B., Schairer C., Mulvihill J.J. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. J. Natl. Cancer Inst. 1989;81:1879–1886. doi: 10.1093/jnci/81.24.1879. [DOI] [PubMed] [Google Scholar]
- 5.Gail M.H., Costantino J.P., Pee D., Bondy M., Newman L., Selvan M., Anderson G.L., Malone K.E., Marchbanks P.A., McCaskill-Stevens W., et al. Projecting individualized absolute invasive breast cancer risk in African American women. J. Natl. Cancer Inst. 2007;99:1782–1792. doi: 10.1093/jnci/djm223. [DOI] [PubMed] [Google Scholar]
- 6.Matsuno R.K., Costantino J.P., Ziegler R.G., Anderson G.L., Li H., Pee D., Gail M.H. Projecting individualized absolute invasive breast cancer risk in Asian and Pacific Islander American women. J. Natl. Cancer Inst. 2011;103:951–961. doi: 10.1093/jnci/djr154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tice J.A., Cummings S.R., Smith-Bindman R., Ichikawa L., Barlow W.E., Kerlikowske K. Using clinical factors and mammographic breast density to estimate breast cancer risk: Development and validation of a new predictive model. Ann. Intern. Med. 2008;148:337–347. doi: 10.7326/0003-4819-148-5-200803040-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sickles E.A., D’Orsi C.J., Bassett L.W., Appleton C.M., Berg W.A., Burnside E.S. ACR BI-RADS® Atlas, Breast Imaging Reporting and Data System. Vol. 5. American College of Radiology; Reston, VA, USA: 2013. ACR BI-RADS® Mammography; pp. 134–136. [Google Scholar]
- 9.Berry D.A., Parmigiani G., Sanchez J., Schildkraut J., Winer E. Probability of carrying a mutation of breast-ovarian cancer gene BRCA1 based on family history. J. Natl. Cancer Inst. 1997;89:227–238. doi: 10.1093/jnci/89.3.227. [DOI] [PubMed] [Google Scholar]
- 10.Guan Z., Huang T., McCarthy A.M., Hughes K.S., Semine A., Uno H., Trippa L., Parmigiani G., Braun D. Combining Breast Cancer Risk Prediction Models. arXiv. 2020 doi: 10.3390/cancers15041090.2008.01019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCarthy A.M., Guan Z., Welch M., Griffin M.E., Sippo D.A., Deng Z., Coopey S.B., Acar A., Semine A., Parmigiani G., et al. Performance of Breast Cancer Risk-Assessment Models in a Large Mammography Cohort. J. Natl. Cancer Inst. 2020;112:489–497. doi: 10.1093/jnci/djz177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howlader N., Altekruse S.F., Li C.I., Chen V.W., Clarke C.A., Ries L.A., Cronin K.A. US incidence of breast cancer subtypes defined by joint hormone receptor and HER2 status. J. Natl. Cancer Inst. 2014;106:dju055. doi: 10.1093/jnci/dju055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phipps A.I., Ichikawa L., Bowles E.J., Carney P.A., Kerlikowske K., Miglioretti D.L., Buist D.S. Defining menopausal status in epidemiologic studies: A comparison of multiple approaches and their effects on breast cancer rates. Maturitas. 2010;67:60–66. doi: 10.1016/j.maturitas.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher B., Costantino J.P., Wickerham D.L., Redmond C.K., Kavanah M., Cronin W.M., Vogel V., Robidoux A., Dimitrov N., Atkins J., et al. Tamoxifen for prevention of breast cancer: Report of the National Surgical Adjuvant Breast and Bowel Project P-1 Study. J. Natl. Cancer Inst. 1998;90:1371–1388. doi: 10.1093/jnci/90.18.1371. [DOI] [PubMed] [Google Scholar]
- 15.Vogel V.G., Costantino J.P., Wickerham D.L., Cronin W.M., Cecchini R.S., Atkins J.N., Bevers T.B., Fehrenbacher L., Pajon E.R., Jr., Wade J.L., 3rd, et al. Effects of tamoxifen vs raloxifene on the risk of developing invasive breast cancer and other disease outcomes: The NSABP Study of Tamoxifen and Raloxifene (STAR) P-2 trial. JAMA. 2006;295:2727–2741. doi: 10.1001/jama.295.23.joc60074. [DOI] [PubMed] [Google Scholar]
- 16.Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.C., Müller M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amir E., Evans D.G., Shenton A., Lalloo F., Moran A., Boggis C., Wilson M., Howell A. Evaluation of breast cancer risk assessment packages in the family history evaluation and screening programme. J. Med. Genet. 2003;40:807–814. doi: 10.1136/jmg.40.11.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anothaisintawee T., Teerawattananon Y., Wiratkapun C., Kasamesup V., Thakkinstian A. Risk prediction models of breast cancer: A systematic review of model performances. Breast Cancer Res. Treat. 2012;133:1–10. doi: 10.1007/s10549-011-1853-z. [DOI] [PubMed] [Google Scholar]
- 19.Brentnall A.R., Cuzick J., Buist D.S.M., Bowles E.J.A. Long-term Accuracy of Breast Cancer Risk Assessment Combining Classic Risk Factors and Breast Density. JAMA Oncol. 2018;4:e180174. doi: 10.1001/jamaoncol.2018.0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brentnall A.R., Harkness E.F., Astley S.M., Donnelly L.S., Stavrinos P., Sampson S., Fox L., Sergeant J.C., Harvie M.N., Wilson M., et al. Mammographic density adds accuracy to both the Tyrer-Cuzick and Gail breast cancer risk models in a prospective UK screening cohort. Breast Cancer Res. 2015;17:147. doi: 10.1186/s13058-015-0653-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chlebowski R.T., Anderson G.L., Lane D.S., Aragaki A.K., Rohan T., Yasmeen S., Sarto G., Rosenberg C.A., Hubbell F.A., Women’s Health Initiative I. Predicting risk of breast cancer in postmenopausal women by hormone receptor status. J. Natl. Cancer Inst. 2007;99:1695–1705. doi: 10.1093/jnci/djm224. [DOI] [PubMed] [Google Scholar]
- 22.Dite G.S., MacInnis R.J., Bickerstaffe A., Dowty J.G., Allman R., Apicella C., Milne R.L., Tsimiklis H., Phillips K.A., Giles G.G., et al. Breast Cancer Risk Prediction Using Clinical Models and 77 Independent Risk-Associated SNPs for Women Aged Under 50 Years: Australian Breast Cancer Family Registry. Cancer Epidemiol. Biomark. Prev. 2016;25:359–365. doi: 10.1158/1055-9965.EPI-15-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Louro J., Posso M., Hilton Boon M., Roman M., Domingo L., Castells X., Sala M. A systematic review and quality assessment of individualised breast cancer risk prediction models. Br. J. Cancer. 2019;121:76–85. doi: 10.1038/s41416-019-0476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McTiernan A., Kuniyuki A., Yasui Y., Bowen D., Burke W., Culver J.B., Anderson R., Durfy S. Comparisons of two breast cancer risk estimates in women with a family history of breast cancer. Cancer Epidemiol. Biomark. Prev. 2001;10:333–338. [PubMed] [Google Scholar]
- 25.Quante A.S., Whittemore A.S., Shriver T., Strauch K., Terry M.B. Breast cancer risk assessment across the risk continuum: Genetic and nongenetic risk factors contributing to differential model performance. Breast Cancer Res. 2012;14:R144. doi: 10.1186/bcr3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rockhill B., Spiegelman D., Byrne C., Hunter D.J., Colditz G.A. Validation of the Gail et al. model of breast cancer risk prediction and implications for chemoprevention. J. Natl. Cancer Inst. 2001;93:358–366. doi: 10.1093/jnci/93.5.358. [DOI] [PubMed] [Google Scholar]
- 27.Terry M.B., Liao Y., Whittemore A.S., Leoce N., Buchsbaum R., Zeinomar N., Dite G.S., Chung W.K., Knight J.A., Southey M.C., et al. 10-year performance of four models of breast cancer risk: A validation study. Lancet Oncol. 2019 doi: 10.1016/S1470-2045(18)30902-1. [DOI] [PubMed] [Google Scholar]
- 28.Tice J.A., Bissell M.C.S., Miglioretti D.L., Gard C.C., Rauscher G.H., Dabbous F.M., Kerlikowske K. Validation of the breast cancer surveillance consortium model of breast cancer risk. Breast Cancer Res. Treat. 2019;175:519–523. doi: 10.1007/s10549-019-05167-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X., Huang Y., Li L., Dai H., Song F., Chen K. Assessment of performance of the Gail model for predicting breast cancer risk: A systematic review and meta-analysis with trial sequential analysis. Breast Cancer Res. 2018;20:18. doi: 10.1186/s13058-018-0947-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zardavas D., Te Marvelde L., Milne R.L., Fumagalli D., Fountzilas G., Kotoula V., Razis E., Papaxoinis G., Joensuu H., Moynahan M.E., et al. Tumor PIK3CA Genotype and Prognosis in Early-Stage Breast Cancer: A Pooled Analysis of Individual Patient Data. J. Clin. Oncol. 2018;36:981–990. doi: 10.1200/JCO.2017.74.8301. [DOI] [PubMed] [Google Scholar]
- 31.Adams-Campbell L.L., Makambi K.H., Palmer J.R., Rosenberg L. Diagnostic accuracy of the Gail model in the Black Women’s Health Study. Breast J. 2007;13:332–336. doi: 10.1111/j.1524-4741.2007.00439.x. [DOI] [PubMed] [Google Scholar]
- 32.Sprague B.L., Conant E.F., Onega T., Garcia M.P., Beaber E.F., Herschorn S.D., Lehman C.D., Tosteson A.N., Lacson R., Schnall M.D., et al. Variation in Mammographic Breast Density Assessments Among Radiologists in Clinical Practice: A Multicenter Observational Study. Ann. Intern. Med. 2016;165:457–464. doi: 10.7326/M15-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Engmann N.J., Scott C.G., Jensen M.R., Winham S., Miglioretti D.L., Ma L., Brandt K., Mahmoudzadeh A., Whaley D.H., Hruska C., et al. Combined effect of volumetric breast density and body mass index on breast cancer risk. Breast Cancer Res. Treat. 2019;177:165–173. doi: 10.1007/s10549-019-05283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kerlikowske K., Walker R., Miglioretti D.L., Desai A., Ballard-Barbash R., Buist D.S. Obesity, mammography use and accuracy, and advanced breast cancer risk. J. Natl. Cancer Inst. 2008;100:1724–1733. doi: 10.1093/jnci/djn388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCarthy A.M., Ehsan S., Appel S., Welch M., He W., Bahl M., Chen J., Lehman C.D., Armstrong K. Risk factors for an advanced breast cancer diagnosis within 2 years of a negative mammogram. Cancer. 2021;127:3334–3342. doi: 10.1002/cncr.33661. [DOI] [PubMed] [Google Scholar]
- 36.Hurson A.N., Pal Choudhury P., Gao C., Husing A., Eriksson M., Shi M., Jones M.E., Evans D.G.R., Milne R.L., Gaudet M.M., et al. Prospective evaluation of a breast-cancer risk model integrating classical risk factors and polygenic risk in 15 cohorts from six countries. Int. J. Epidemiol. 2021 doi: 10.1093/ije/dyab036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying this article cannot be shared publicly in order to protect patient privacy. The data may be shared in a de-identified format on reasonable request to the corresponding author.