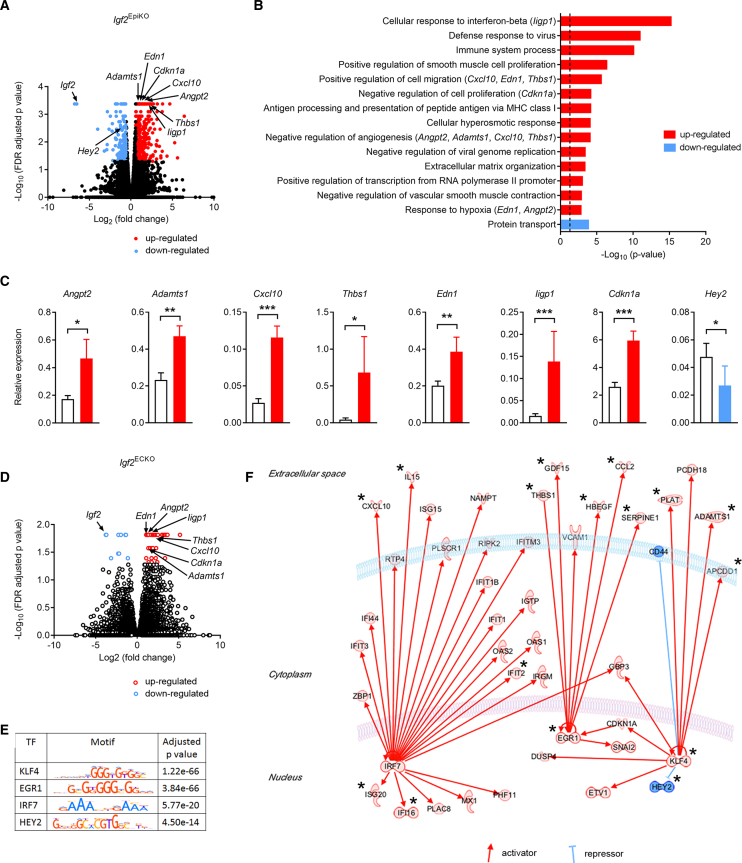
Figure 5.
IGF2 signaling regulates angiogenic properties of endothelial cells
(A) Volcano plot representation of DEGs identified by RNA-seq in E16 FPEC (Igf2EpiKO versus controls). Significant upregulated and downregulated DEGs (false discovery rate [FDR] < 0.05) are shown with red and blue, respectively.
(B) Top scoring biological processes enriched in DEGs. Biologically validated DEGS are listed in parentheses. The dotted line corresponds to FDR-corrected p value of 0.05.
(C) Biological validation. Data are shown as averages (n = 11–12 samples per group); error bars are SEM; ∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001 calculated by Mann-Whitney tests.
(D) Volcano plot representation of DEGs identified by RNA-seq in E16 FPEC (Igf2ECKO versus controls). Significant upregulated and downregulated DEGs (FDR < 0.05) are shown with red and blue, respectively.
(E) Transcription factors (TFs) identified by analysis of motif enrichment (AME).
(F) IPA regulatory network built with the four TFs identified using AME analysis. Proteins labeled with a star are known regulators of angiogenesis (angiostatic or pro-angiogenic factors) and key references are listed in Table S4. See also Figures S5C and S5D and Tables S3 and S4.