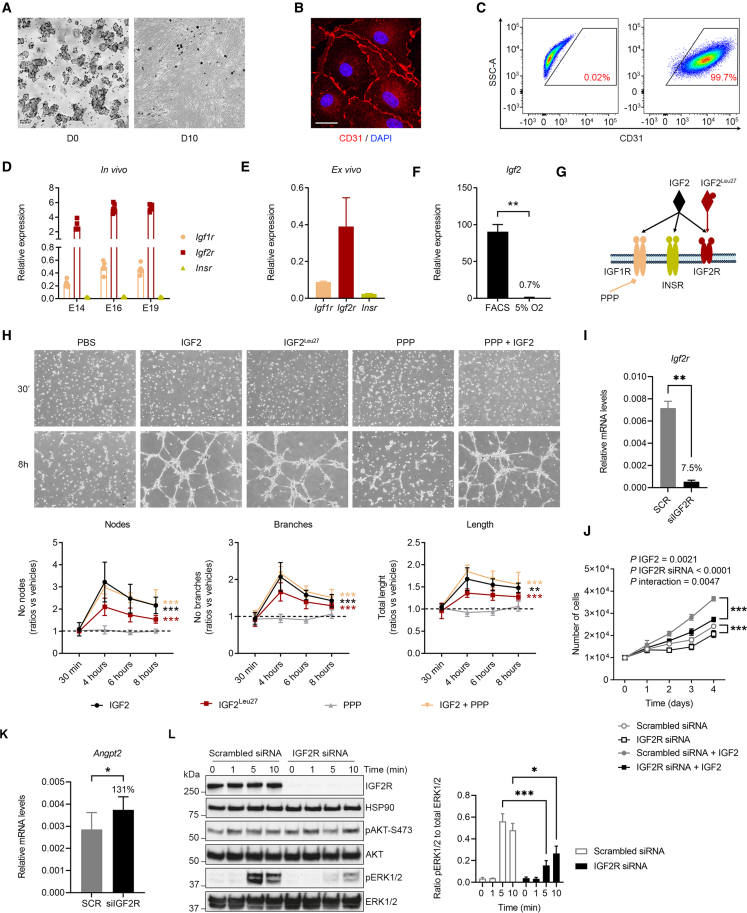
Figure 6.
IGF2 Acts on FPECs via IGF2R-ERK signaling ex vivo
(A) Primary FPEC isolated from E16 Lz: D0—freshly isolated cells; D10—FPEC at passage one (P1, 10 days of culture).
(B) Confocal imaging of passage one FPEC, stained for CD31 (scale bars, 20 μm).
(C) Flow cytometry analysis of P1 FPEC stained for CD31, demonstrating that these are almost exclusively CD31+.
(D) qRT-PCR analysis for Igf1r, Igf2r, and Insr in FPECs isolated by FACS (n = 6–7 per group).
(E) Relative expression of the three IGF receptors in P1 FPEC.
(F) qRT-PCR analysis of Igf2 mRNA levels in P1 FPEC cultured in 5% O2 versus primary FPEC isolated from E16 Lz by FACS.
(G) Schematic representation of IGF2 and IGF receptors. IGF2Leu27 analog acts specifically on IGF2R and picropodophyllin (PPP) inhibits phosphorylation of IGF1R.
(H) Representative images of capillary-like tube formation assay in primary FPEC seeded on matrigel and exposed to exogenous IGF2, IGF2Leu27, PPP, or PPP+IGF2 (equal seeding of cell numbers at 30 min and tube formation at 8 h), and quantification of number of nodes, branches, and total length (n = 5–6 independent experiments).
(I) qRT-PCR analysis of Igf2r mRNA levels in primary FPECs upon knockdown by siRNA (n = 8 samples/group).
(J) Proliferation assay of primary FPEC with or without IGF2R siRNA knockdown, in presence or absence of IGF2, on 4 consecutive days after plating. Cells with IGF2R siRNA knockdown exhibit significant proliferation defects that are further accentuated upon IGF2 treatment (n = 5 biological replicates per group).
(K) qRT-PCR analysis of Angpt2 mRNA levels in primary FPECs transfected with scrambled siRNA or IGF2R siRNA, upon 4 days of treatment with 50 ng/mL mouse recombinant IGF2 (n = 8 samples/group).
(L) Left side: identification of delayed ERK1/2 phosphorylation in FPECs with IGF2R siRNA knockdown upon acute treatment with 50 ng/mL mouse recombinant IGF2. HSP90 was used as internal control for protein loading. Right side: quantification of ratios pERK1/2 to total ERK1/2 for n = 3 independent biological replicates. For all graphs, data are presented as averages or individual values and error bars represent SEM. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 calculated by a Mann-Whitney test in (F), two-way ANOVA tests with Sidak’s multiple comparisons test in (H), (J), and (L), Wilcoxon matched-pairs signed rank test in (I) and paired Student’s t test in (K).