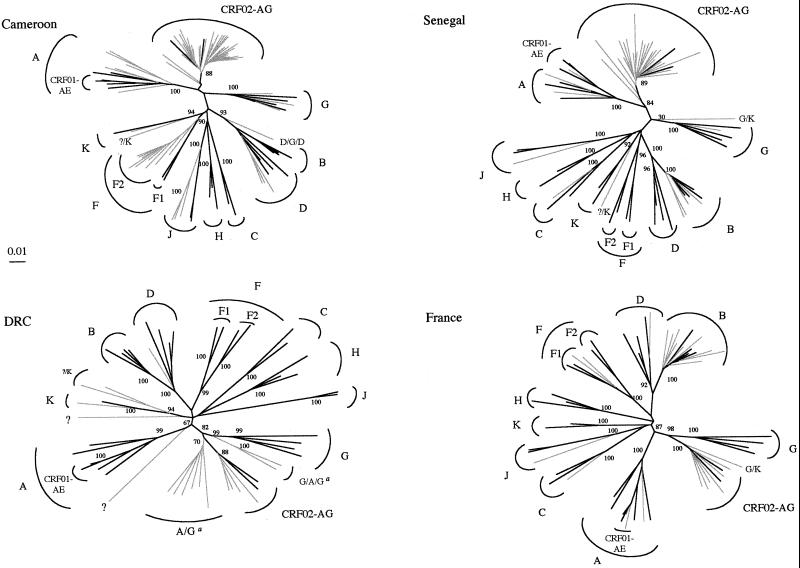
FIG. 1.
Unrooted phylogenetic trees of protease and RT nucleotide sequences (1,600 bp) from 138 group M isolates (in gray) and from reference strains (in black) representing the different subtypes. The trees were generated as described in Materials and Methods. The reference sequences (8) used in the trees were as follows: A.U455, A.Q2317, A.92UG037.1, B.OYI, B.HXB2, B.JRFL, B.RF, D.NDK, D.ELI, D.84ZR085, D.94UG114.1, C.ETH2220, C.92BR025.8, F1.93BR020.1, F1.FIN9363, G.SE6165, G.92NG083, G(A).92NG003, G.HH8793.1.1, H.VI991, H.VI997, H.90CF056.1, J.SE9173.3, J.SE9280.9, CRF01-AE.CM240, CRF01-AE.93TH253.3, CRF01-AE.90CF402.1, CRF02-AG.IBNG, and CRF02-AG.DJ263. The reference sequences for the K and F2 subtypes (41, 42) were used either as references or as samples (K.96CAM.MP535, K.97ZR.EQTB11, F2.95CM.MP255, F2.95CM.MP257). For sequences that did not cluster with high bootstrap values with any of the known subtypes or CRFs, the results of complementary analyses indicating their recombinant nature are shown. a, the isolates designated A/G and G/A/G are intersubtype A/G recombinant viruses in which the breakpoints between A and G are different from those for the prototype CRF02-AG strain. The numbers at the branch points indicate bootstrap values as percentages.